diff --git a/docs/zh/examples/tadf.md b/docs/zh/examples/tadf.md
new file mode 100644
index 0000000000..34304a5f03
--- /dev/null
+++ b/docs/zh/examples/tadf.md
@@ -0,0 +1,176 @@
+# 热活化延迟荧光材料(TADF)性质预测
+
+!!! note
+
+ 1. 开始训练、评估前,请先确保性质数据文件(.dat)和SMILES(smis.txt)数据文件的存在,并对应修改 yaml 配置文件中的 `data_dir` 为性质数据文件路径,`sim_dir` 为SMILES数据文件路径。
+ 2. 开始训练、评估前,请安装 `rdkit` 等,相关依赖请执行`pip install -r requirements.txt`安装。
+
+| 预训练模型 | 指标 |
+|:--| :--|
+| [Est.pdparams](https://paddle-org.bj.bcebos.com/paddlescience/models/TADF/Est/Est_model.pdparams) | loss(MAE): 0.045 |
+| [f.pdparams](https://paddle-org.bj.bcebos.com/paddlescience/models/TADF/f/f_model.pdparams) | loss(MAE): 0.036 |
+| [angle.pdparams](https://paddle-org.bj.bcebos.com/paddlescience/models/TADF/angle/angle_model.pdparams) | loss(MAE): 0.041 |
+
+=== "模型训练命令"
+
+ ``` sh
+ # Est 预测:
+ cd TADF_Est
+ wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/Est/Est.dat https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/smis.txt
+ python Est.py mode=train
+
+ #f 预测:
+ cd TADF_f
+ wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/f/f.dat https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/smis.txt
+ python f.py mode=train
+
+ # angle 预测:
+ cd TADF_angle
+ wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/angle/angle.dat https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/smis.txt
+ python angle.py mode=train
+ ```
+
+=== "模型评估命令"
+
+ ``` sh
+ # Est 评估:
+ cd TADF_Est
+ wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/Est/Est.dat \ https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/smis.txt
+ python Est.py mode=eval EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/TADF/Est/Est_model.pdparams
+
+ # f 评估:
+ cd TADF_f
+ wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/f/f.dat \ https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/smis.txt
+ python f.py mode=eval EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/TADF/f/f_model.pdparams
+
+ # angle 评估:
+ cd TADF_angle
+ wget -nc https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/f/f.dat \ https://paddle-org.bj.bcebos.com/paddlescience/datasets/TADF/smis.txt
+ python angle.py mode=eval EVAL.pretrained_model_path=https://paddle-org.bj.bcebos.com/paddlescience/models/TADF/angle/angle_model.pdparams
+ ```
+
+## 1. 背景简介
+
+有机发光二极管(OLED)具有高效率、结构灵活和低成本的优势,在先进显示和照明技术中受到广泛关注。在有机发光二极管器件中,电注入载流子以1:3的比例形成单线态和三线态激子。以纯荧光材料为发光材料构建的OLED发光效率IQE理论极限为25%。另一方面,有机金属复合物发光材料通过引入稀有金属(Ir,Pt等)带来强自旋轨道耦合(SOC),可以将单线态激子通过系间窜越过程转化成三线态激子,从而利用三线态激子发出磷光,其IQE可达100%,但是稀有金属价格昂贵,为推广使用带来了阻碍。热活化延迟荧光材料(TADF)为解决这些问题提供了新思路,并引起了广泛关注。在TADF中,三线态通过逆系间窜越过程(RISC)转化成单重态并发出荧光,从而实现100%的IQE,而RISC过程很大程度上取决于最低单线态(S1)和最低三线态(T1) 之间的能隙( $\Delta Est$ )。根据量子力学理论,ΔEST相当于HOMO和LUMO之间的交换积分的两倍。因此TADF分子的常见设计策略是将电子供体(D)和电子受体(A)以明显扭曲的二面角结合以实现HOMO和LUMO在空间上明显的分离。然而,与 $\Delta Est$ 相反,振子强度( $f$ )需要较大的HOMO和LUMO之间的重叠积分,这二者之间的矛盾需要进一步平衡。
+
+## 2. 模型原理
+
+通过高通量计算构建数据集,通过分子结构输入、指纹特征提取、特征降维三个环节实现分子特征表征,随后通过多层非线性变换学习分子结构特征与TADF关键参数间的复杂映射关系,最终实现端到端的性质预测。
+
+## 3. TADF性质预测模型的实现
+
+本样例包括对化学分子的能隙( $\Delta Est$ ),振子强度( $f$ ),电子供体与电子受体间的二面角( $angle$ )三项性质的预测,接下来将以二面角 $angle$ 为例,开始讲解如何基于PaddleScience代码,实现对于TADF性质预测模型的构建、训练、测试和评估。案例的目录结构如下:
+
+``` log
+tadf/
+├──TADF_angle/
+│ ├── config/
+│ │ └── angle.yaml
+│ ├── angle_model.py
+│ ├── angle.dat
+│ ├── angle.py
+│ └── smis.txt
+├── TADF_Est/
+│ └── ...
+├── TADF_f/
+│ └── ...
+└── requirements.txt
+```
+
+### 3.1 数据集准备
+
+我们选择常用的49个受体和50个受体以单键相连的方式进行组合,通过穷举所有可能的组合位点我们得到了44470个分子。通过MMFF94力场优化得到分子的初始结构。从44470个分子中随机提取5136个分子,在B3LYP/6-31G(d)水平下对5136个分子进行基态结构优化,采用TDDFT方法在基态构型下进行激发态性质计算。
+
+本案例所用数据包括性质数据文件(例如angle.dat)和SMILES数据文件(smis.txt)。分子性质数据文件的每一行为一条分子性质;smis.txt的每一行为一个分子的 SMILES描述,即用一串字符把分子结构编码成线性字符串,以第一条数据数据为例
+
+```
+c1cc(-c2c3ccoc3cc3ccoc23)n[nH]1
+```
+其中小写 `c` 代表芳香碳,`n` 代表芳香氮,`[nH]` 代表带一个氢原子的芳香氮。数字 `1`、`2`、`3` 表示环的开闭标记:第一个 `1` 开启了一个环,遇到下一个 `1` 就闭合它。
+
+在依据配置文件的信息逐行对性质数据文件和SMILES数据文件进行加载后,首先通过 `rdkit.Chem.rdFingerprintGenerator` 将分子的SMILES描述转换为 Morgan 指纹。Morgan指纹是一种分子结构的向量化描述,通过局部拓扑被编码为 hash 值,映射到2048位指纹位上。随后,使用PCA把2048维降到主成分保留99%方差的维度。用PaddleScience代码表示如下
+
+``` py linenums="20" title="examples/tadf/TADF_angle/angle_model.py"
+--8<--
+examples/tadf/TADF_angle/angle_model.py:20:66
+--8<--
+```
+
+### 3.2 约束构建
+
+本研究采用监督学习,按照 PaddleScience 的API结构说明,采用内置的 `SupervisedConstraint` 构建监督约束。用 PaddleScience 代码表示如下
+
+``` py linenums="97" title="examples/tadf/TADF_angle/angle_model.py"
+--8<--
+examples/tadf/TADF_angle/angle_model.py:97:108
+--8<--
+```
+
+`SupervisedConstraint` 的第二个参数表示采用均方误差 `MSELoss` 作为损失函数,第三个参数表示约束条件的名字,方便后续对其索引。
+
+### 3.3 模型构建
+
+对于三个预测对象,设计了相同的深度神经网络,网络结构为含有两层隐藏层的神经网络,第一层隐藏层含有587个神经元,第二层隐藏层含有256个神经元,隐藏层之间加入Dropout。以 $angle$ 预测为例,用 PaddleScience 代码表示如下
+
+``` py linenums="110" title="examples/tadf/TADF_angle/angle_model.py"
+--8<--
+examples/tadf/TADF_angle/angle_model.py:110:119
+--8<--
+```
+
+### 3.4 优化器构建
+
+在本案例的angle性质预测中,训练器采用Adam优化器,学习率设置为0.01,`weight_decay` 设置为 1e-5,用 PaddleScience 代码表示如下
+
+``` py linenums="120" title="examples/tadf/TADF_angle/angle_model.py"
+--8<--
+examples/tadf/TADF_angle/angle_model.py:120:125
+--8<--
+```
+
+### 3.5 模型训练
+
+完成上述设置之后,只需要将上述实例化的对象按顺序传递给`ppsci.solver.Solver`,然后启动训练即可。用PaddleScience 代码表示如下
+
+``` py linenums="127" title="examples/tadf/TADF_angle/angle_model.py"
+--8<--
+examples/tadf/TADF_angle/angle_model.py:127:137
+--8<--
+```
+
+## 4. 完整代码
+
+``` py linenums="1" title="examples/tadf/TADF_angle/angle_model.py"
+--8<--
+examples/tadf/TADF_angle/angle_model.py
+--8<--
+```
+
+``` py linenums="1" title="examples/tadf/TADF_angle/angle.py"
+--8<--
+examples/tadf/TADF_angle/angle.py
+--8<--
+```
+
+## 5. 结果展示
+
+下图展示能隙( $\Delta Est$ ),振子强度( $f$ ),电子供体与电子受体间的二面角( $angle$ )三项性质的模型预测结果。
+
+
+ 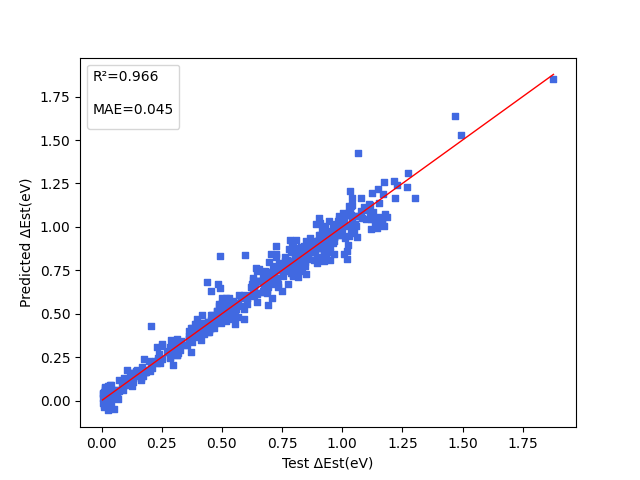{ loading=lazy }
+ 能隙(ΔEst)的模型预测结果
+
+
+
+ 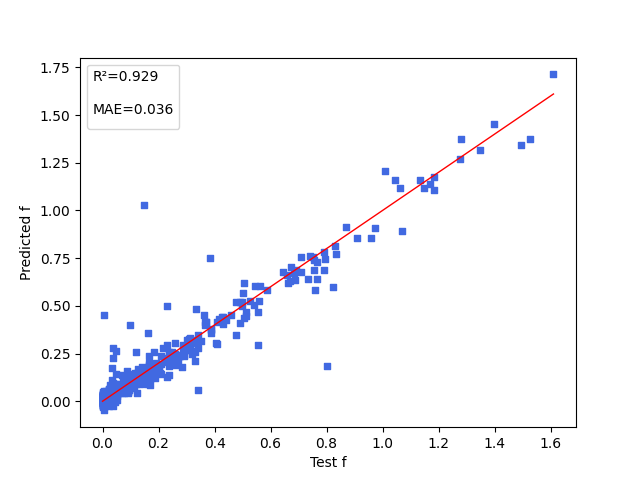{ loading=lazy }
+ 振子强度(f)的模型预测结果
+
+
+
+ 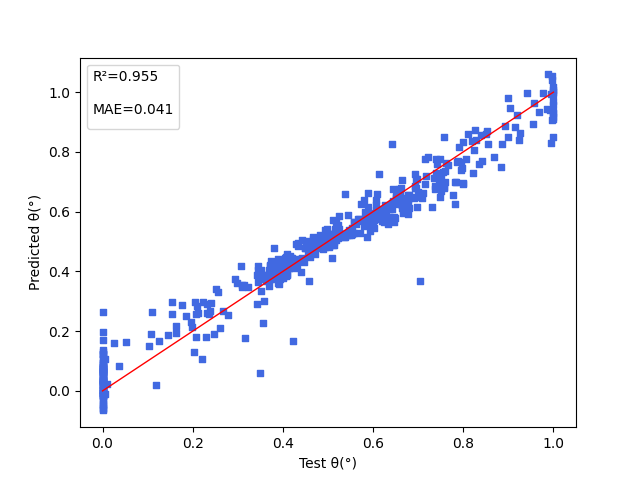{ loading=lazy }
+ 电子供体与电子受体间的二面角(angle)的模型预测结果
+
+
+## 6. 参考文献
+
+[1] Yufei Bu, Qian Peng*, Designing Promising Thermally Activated Delayed Fluroscence Emitters via Machine Learning-Assisted High-Throughput Virtual Screening. J. Phys. Chem. C. 2023. DOI: 10.1021/acs.jpcc.3c05337.
diff --git a/examples/tadf/TADF_Est/Est.py b/examples/tadf/TADF_Est/Est.py
new file mode 100644
index 0000000000..5aa7df1cfb
--- /dev/null
+++ b/examples/tadf/TADF_Est/Est.py
@@ -0,0 +1,22 @@
+import hydra
+from Est_model import evaluate
+from Est_model import featurize_molecules
+from Est_model import load_data
+from Est_model import train
+from omegaconf import DictConfig
+
+
+@hydra.main(version_base=None, config_path="./config", config_name="est.yaml")
+def main(cfg: DictConfig):
+ data, smis = load_data(cfg)
+ X = featurize_molecules(smis)
+ if cfg.mode == "train":
+ train(cfg, X, data)
+ elif cfg.mode == "eval":
+ evaluate(cfg, X, data)
+ else:
+ raise ValueError(f"cfg.mode should in ['train', 'eval'], but got '{cfg.mode}'")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/tadf/TADF_Est/Est_model.py b/examples/tadf/TADF_Est/Est_model.py
new file mode 100644
index 0000000000..e1936c928e
--- /dev/null
+++ b/examples/tadf/TADF_Est/Est_model.py
@@ -0,0 +1,218 @@
+import os
+
+import matplotlib.pyplot as plt
+import numpy as np
+import paddle
+import rdkit.Chem as Chem
+from omegaconf import DictConfig
+from rdkit.Chem import rdFingerprintGenerator
+from sklearn.decomposition import PCA
+from sklearn.model_selection import train_test_split
+
+import ppsci
+
+os.environ["HYDRA_FULL_ERROR"] = "1"
+os.environ["KMP_DUPLICATE_LIB_OK"] = "True"
+plt.rcParams["axes.unicode_minus"] = False
+plt.rcParams["font.sans-serif"] = ["DejaVu Sans"]
+
+# Data preparation
+def load_data(cfg):
+ data_dir = cfg.data_dir
+ sim_dir = cfg.sim_dir
+ angle_dat_path = os.path.join(data_dir)
+ smis_txt_path = os.path.join(sim_dir)
+
+ data = []
+ with open(angle_dat_path) as f:
+ for line in f:
+ num = float(line.strip())
+ data.append(num)
+
+ smis = []
+ with open(smis_txt_path) as f:
+ for line in f:
+ smis.append(line.strip())
+
+ return data, smis
+
+
+def featurize_molecules(smis):
+ vectors = []
+ del_mol = []
+ for s in smis:
+ mol = Chem.MolFromSmiles(s)
+ try:
+ generator = rdFingerprintGenerator.GetMorganGenerator(radius=2, fpSize=2048)
+ fp = generator.GetFingerprint(mol)
+ _input = np.array(list(map(int, fp.ToBitString())))
+ vectors.append(_input)
+ except Exception as e:
+ print(f"Error processing {s}: {e}")
+ del_mol.append(s)
+ pca = PCA(n_components=0.99)
+ pca.fit(vectors)
+ X = pca.transform(vectors)
+ return paddle.to_tensor(X, dtype="float32")
+
+
+def train(cfg: DictConfig, X, data):
+ # k-fold cross validation splitter
+ def k_fold(k, i, X, Y):
+ fold_size = tuple(X.shape)[0] // k
+ val_start = i * fold_size
+ if i != k - 1:
+ val_end = (i + 1) * fold_size
+ x_val, y_val = X[val_start:val_end], Y[val_start:val_end]
+ x_train = np.concatenate((X[0:val_start], X[val_end:]), axis=0)
+ y_train = np.concatenate((Y[0:val_start], Y[val_end:]), axis=0)
+ else:
+ x_val, y_val = X[val_start:], Y[val_start:]
+ x_train = X[0:val_start]
+ y_train = Y[0:val_start]
+ return x_train, y_train, x_val, y_val
+
+ Y = paddle.to_tensor(data, dtype="float32")
+ x_train, y_train, x_test, y_test = k_fold(cfg.TRAIN.k, cfg.TRAIN.i, X, Y)
+ # Prepare feature dictionary
+ x_train = paddle.to_tensor(x_train, dtype="float32")
+ x = {
+ f"key_{i}": paddle.unsqueeze(
+ paddle.to_tensor(x_train[:, i], dtype="float32"), axis=1
+ )
+ for i in range(x_train.shape[1])
+ }
+ y_train = paddle.unsqueeze(paddle.to_tensor(y_train, dtype="float32"), axis=1)
+
+ # Build supervised constraint
+ sup = ppsci.constraint.SupervisedConstraint(
+ dataloader_cfg={
+ "dataset": {
+ "name": "IterableNamedArrayDataset",
+ "input": x,
+ "label": {"u": y_train},
+ },
+ "batch_size": cfg.TRAIN.batch_size,
+ },
+ loss=ppsci.loss.MSELoss("mean"),
+ name="sup",
+ )
+
+ # Set model architecture parameters
+ hidden_size = [587, 256]
+ num_layers = None
+ # Instantiate TADF model
+ model = ppsci.arch.TADF(
+ input_keys=tuple(x.keys()),
+ hidden_size=hidden_size,
+ num_layers=num_layers,
+ **cfg.MODEL,
+ )
+ optimizer = ppsci.optimizer.optimizer.Adam(
+ cfg.TRAIN.learning_rate,
+ beta1=0.9,
+ beta2=0.99,
+ weight_decay=cfg.TRAIN.weight_decay,
+ )(model)
+
+ # Build solver for training
+ solver = ppsci.solver.Solver(
+ model,
+ constraint={sup.name: sup},
+ optimizer=optimizer,
+ epochs=cfg.TRAIN.epochs,
+ eval_during_train=False,
+ iters_per_epoch=cfg.TRAIN.iters_per_epoch,
+ )
+ try:
+ solver.train()
+ except Exception as ex:
+ print("error", ex)
+
+
+def evaluate(cfg: DictConfig, X, data):
+ y_full = paddle.to_tensor(data, dtype="float32")
+ X_np = X.numpy()
+ y_np = y_full.numpy()
+ X_train_np, X_test_np, y_train_np, y_test_np = train_test_split(
+ X_np,
+ y_np,
+ test_size=cfg.EVAL.test_size,
+ random_state=cfg.EVAL.seed,
+ )
+ x_test = paddle.to_tensor(X_test_np, dtype="float32")
+ y_test = paddle.to_tensor(y_test_np, dtype="float32")
+
+ x_dict = {
+ f"key_{i}": paddle.unsqueeze(x_test[:, i], axis=1)
+ for i in range(x_test.shape[1])
+ }
+
+ test_validator = ppsci.validate.SupervisedValidator(
+ dataloader_cfg={
+ "dataset": {
+ "name": "IterableNamedArrayDataset",
+ "input": x_dict,
+ "label": {"u": paddle.unsqueeze(y_test, axis=1)},
+ },
+ "batch_size": cfg.EVAL.batch_size,
+ "shuffle": False,
+ },
+ loss=ppsci.loss.MSELoss("mean"),
+ metric={
+ "MAE": ppsci.metric.MAE(),
+ "RMSE": ppsci.metric.RMSE(),
+ "R2": ppsci.metric.R2Score(),
+ },
+ name="test_eval",
+ )
+ validators = {"test_eval": test_validator}
+
+ model = ppsci.arch.TADF(
+ input_keys=tuple(x_dict.keys()),
+ hidden_size=[587, 256],
+ num_layers=None,
+ **cfg.MODEL,
+ )
+
+ solver = ppsci.solver.Solver(
+ model,
+ validator=validators,
+ cfg=cfg,
+ )
+
+ _, metric_dict = solver.eval()
+
+ ypred = model(x_dict)["u"].numpy()
+ ytrue = paddle.unsqueeze(y_test, axis=1).numpy()
+
+ mae = metric_dict["MAE"]["u"]
+ rmse = metric_dict["RMSE"]["u"]
+ r2 = metric_dict["R2"]["u"]
+
+ print("Evaluation metrics:")
+ print(f"MAE: {mae:.4f}")
+ print(f"RMSE: {rmse:.4f}")
+ print(f"R2: {r2:.4f}")
+
+ plt.scatter(
+ ytrue,
+ ypred,
+ s=15,
+ color="royalblue",
+ marker="s",
+ linewidth=1,
+ )
+ plt.plot(
+ [ytrue.min(), ytrue.max()],
+ [ytrue.min(), ytrue.max()],
+ "r-",
+ lw=1,
+ )
+ plt.legend(title=f"R²={r2:.3f}\n\nMAE={mae:.3f}")
+ plt.xlabel("Test θ(°)")
+ plt.ylabel("Predicted θ(°)")
+ save_path = "test_est.png"
+ plt.savefig(save_path)
+ print(f"图片已保存至:{save_path}")
+ plt.show()
diff --git a/examples/tadf/TADF_Est/config/est.yaml b/examples/tadf/TADF_Est/config/est.yaml
new file mode 100644
index 0000000000..50b7bf6a02
--- /dev/null
+++ b/examples/tadf/TADF_Est/config/est.yaml
@@ -0,0 +1,61 @@
+defaults:
+ - ppsci_default
+ - TRAIN: train_default
+ - TRAIN/ema: ema_default
+ - TRAIN/swa: swa_default
+ - EVAL: eval_default
+ - INFER: infer_default
+ - hydra/job/config/override_dirname/exclude_keys: exclude_keys_default
+ - _self_
+
+hydra:
+ run:
+ # dynamic output directory according to running time and override name
+ dir: outputs_Est/${now:%Y-%m-%d}/${now:%H-%M-%S}/${hydra.job.override_dirname}
+ job:
+ name: ${mode} # name of logfile
+ chdir: false # keep current working directory unchanged
+ callbacks:
+ init_callback:
+ _target_: ppsci.utils.callbacks.InitCallback
+ sweep:
+ # output directory for multirun
+ dir: ${hydra.run.dir}
+ subdir: ./
+
+# general settings
+mode: eval # running mode: train/eval
+seed: 42
+output_dir: ${hydra:run.dir}
+log_freq: 20
+use_tbd: false
+VIV_DATA_PATH: "./Est.dat"
+data_dir: "./Est.dat"
+sim_dir: "./smis.txt"
+
+# model settings
+MODEL:
+ output_keys: ["u"]
+ activation: "relu"
+ dropout: 0.5
+
+# training settings
+TRAIN:
+ epochs: 200
+ iters_per_epoch: 20
+ save_freq: 100
+ eval_during_train: False
+ batch_size: 8
+ learning_rate: 0.0001
+ weight_decay: 1e-5
+ pretrained_model_path: null
+ checkpoint_path: null
+ k: 9
+ i: 2
+
+# evaluation settings
+EVAL:
+ test_size: 0.1
+ pretrained_model_path: null
+ batch_size: 8
+ seed: 20
diff --git a/examples/tadf/TADF_angle/angle.py b/examples/tadf/TADF_angle/angle.py
new file mode 100644
index 0000000000..020087f585
--- /dev/null
+++ b/examples/tadf/TADF_angle/angle.py
@@ -0,0 +1,22 @@
+import hydra
+from angle_model import evaluate
+from angle_model import featurize_molecules
+from angle_model import load_data
+from angle_model import train
+from omegaconf import DictConfig
+
+
+@hydra.main(version_base=None, config_path="./config", config_name="angle.yaml")
+def main(cfg: DictConfig):
+ data, smis = load_data(cfg)
+ X = featurize_molecules(smis)
+ if cfg.mode == "train":
+ train(cfg, X, data)
+ elif cfg.mode == "eval":
+ evaluate(cfg, X, data)
+ else:
+ raise ValueError(f"cfg.mode should be 'train' or 'eval', but got '{cfg.mode}'")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/tadf/TADF_angle/angle_model.py b/examples/tadf/TADF_angle/angle_model.py
new file mode 100644
index 0000000000..4947e522a7
--- /dev/null
+++ b/examples/tadf/TADF_angle/angle_model.py
@@ -0,0 +1,226 @@
+import os
+
+import matplotlib.pyplot as plt
+import numpy as np
+import paddle
+import rdkit.Chem as Chem
+from omegaconf import DictConfig
+from rdkit.Chem import rdFingerprintGenerator
+from sklearn.decomposition import PCA
+from sklearn.model_selection import train_test_split
+
+import ppsci
+
+paddle.set_device("gpu:0")
+os.environ["HYDRA_FULL_ERROR"] = "1"
+os.environ["KMP_DUPLICATE_LIB_OK"] = "True"
+plt.rcParams["axes.unicode_minus"] = False
+plt.rcParams["font.sans-serif"] = ["DejaVu Sans"]
+# Data preparation
+def load_data(cfg):
+ data_dir = cfg.data_dir
+ sim_dir = cfg.sim_dir
+ angle_dat_path = os.path.join(data_dir)
+ smis_txt_path = os.path.join(sim_dir)
+
+ data = []
+ with open(angle_dat_path) as f:
+ for line in f:
+ num = float(line.strip()) / 90
+ data.append(num)
+
+ smis = []
+ with open(smis_txt_path) as f:
+ for line in f:
+ smis.append(line.strip())
+
+ return data, smis
+
+
+def featurize_molecules(smis):
+ vectors = []
+ del_mol = []
+ for s in smis:
+ # Convert SMILES to RDKit molecule object
+ mol = Chem.MolFromSmiles(s)
+ try:
+ # Create Morgan fingerprint generator (radius 2, 2048-bit vector)
+ generator = rdFingerprintGenerator.GetMorganGenerator(radius=2, fpSize=2048)
+
+ # Generate fingerprint for current molecule
+ fp = generator.GetFingerprint(mol)
+
+ _input = np.array(list(map(int, fp.ToBitString())))
+ vectors.append(_input)
+ except Exception as e:
+ print(f"Error processing {s}: {e}")
+ del_mol.append(s)
+
+ # Initialize PCA preserving 99% of variance
+ pca = PCA(n_components=0.99)
+
+ # Learn PCA transformation from fingerprint vectors
+ pca.fit(vectors)
+ X = pca.transform(vectors)
+ return paddle.to_tensor(X, dtype="float32")
+
+
+def train(cfg: DictConfig, X, data):
+ # k-fold cross validation splitter
+ def k_fold(k, i, X, Y):
+ fold_size = X.shape[0] // k
+ val_start = i * fold_size
+ if i != k - 1:
+ val_end = (i + 1) * fold_size
+ x_val, y_val = X[val_start:val_end], Y[val_start:val_end]
+ x_train = paddle.concat((X[0:val_start], X[val_end:]), axis=0)
+ y_train = paddle.concat((Y[0:val_start], Y[val_end:]), axis=0)
+ else:
+ x_val, y_val = X[val_start:], Y[val_start:]
+ x_train = X[0:val_start]
+ y_train = Y[0:val_start]
+ return x_train, y_train, x_val, y_val
+
+ Y = paddle.to_tensor(data, dtype="float32")
+ x_train, y_train, x_test, y_test = k_fold(cfg.TRAIN.k, cfg.TRAIN.i, X, Y)
+ # Prepare feature dictionary
+ x = {
+ f"key_{i}": paddle.unsqueeze(
+ paddle.to_tensor(x_train[:, i], dtype="float32"), axis=1
+ )
+ for i in range(x_train.shape[1])
+ }
+ y_train = paddle.unsqueeze(paddle.to_tensor(y_train, dtype="float32"), axis=1)
+
+ # Build supervised constraint
+ sup = ppsci.constraint.SupervisedConstraint(
+ dataloader_cfg={
+ "dataset": {
+ "name": "IterableNamedArrayDataset",
+ "input": x,
+ "label": {"u": y_train},
+ },
+ "batch_size": cfg.TRAIN.batch_size,
+ },
+ loss=ppsci.loss.MSELoss("mean"),
+ name="sup",
+ )
+
+ # Set model architecture parameters
+ hidden_size = [587, 256]
+ num_layers = None
+ # Instantiate TADF model
+ model = ppsci.arch.TADF(
+ input_keys=tuple(x.keys()),
+ hidden_size=hidden_size,
+ num_layers=num_layers,
+ **cfg.MODEL,
+ )
+ optimizer = ppsci.optimizer.Adam(
+ learning_rate=cfg.TRAIN.learning_rate,
+ beta1=0.9,
+ beta2=0.99,
+ weight_decay=cfg.TRAIN.weight_decay,
+ )(model)
+
+ # Build solver for training
+ solver = ppsci.solver.Solver(
+ model,
+ constraint={sup.name: sup},
+ optimizer=optimizer,
+ epochs=cfg.TRAIN.epochs,
+ eval_during_train=False,
+ iters_per_epoch=cfg.TRAIN.iters_per_epoch,
+ )
+ try:
+ solver.train()
+ except Exception as ex:
+ print(ex)
+
+
+def evaluate(cfg: DictConfig, X, data):
+ y_full = paddle.to_tensor(data, dtype="float32")
+ X_np = X.numpy()
+ y_np = y_full.numpy()
+ X_train_np, X_test_np, y_train_np, y_test_np = train_test_split(
+ X_np,
+ y_np,
+ test_size=cfg.EVAL.test_size,
+ random_state=cfg.EVAL.seed,
+ )
+ x_test = paddle.to_tensor(X_test_np, dtype="float32")
+ y_test = paddle.to_tensor(y_test_np, dtype="float32")
+
+ x_dict = {
+ f"key_{i}": paddle.unsqueeze(x_test[:, i], axis=1)
+ for i in range(x_test.shape[1])
+ }
+
+ test_validator = ppsci.validate.SupervisedValidator(
+ dataloader_cfg={
+ "dataset": {
+ "name": "IterableNamedArrayDataset",
+ "input": x_dict,
+ "label": {"u": paddle.unsqueeze(y_test, axis=1)},
+ },
+ "batch_size": cfg.EVAL.batch_size,
+ "shuffle": False,
+ },
+ loss=ppsci.loss.MSELoss("mean"),
+ metric={
+ "MAE": ppsci.metric.MAE(),
+ "RMSE": ppsci.metric.RMSE(),
+ "R2": ppsci.metric.R2Score(),
+ },
+ name="test_eval",
+ )
+ validators = {"test_eval": test_validator}
+
+ model = ppsci.arch.TADF(
+ input_keys=tuple(x_dict.keys()),
+ hidden_size=[587, 256],
+ num_layers=None,
+ **cfg.MODEL,
+ )
+
+ solver = ppsci.solver.Solver(
+ model,
+ validator=validators,
+ cfg=cfg,
+ )
+
+ _, metric_dict = solver.eval()
+
+ ypred = model(x_dict)["u"].numpy()
+ ytrue = paddle.unsqueeze(y_test, axis=1).numpy()
+
+ mae = metric_dict["MAE"]["u"]
+ rmse = metric_dict["RMSE"]["u"]
+ r2 = metric_dict["R2"]["u"]
+
+ print("Evaluation metrics:")
+ print(f"MAE: {mae:.4f}")
+ print(f"RMSE: {rmse:.4f}")
+ print(f"R2: {r2:.4f}")
+
+ plt.scatter(
+ ytrue,
+ ypred,
+ s=15,
+ color="royalblue",
+ marker="s",
+ linewidth=1,
+ )
+ plt.plot(
+ [ytrue.min(), ytrue.max()],
+ [ytrue.min(), ytrue.max()],
+ "r-",
+ lw=1,
+ )
+ plt.legend(title=f"R²={r2:.3f}\n\nMAE={mae:.3f}")
+ plt.xlabel("Test θ(°)")
+ plt.ylabel("Predicted θ(°)")
+ save_path = "test_angle.png"
+ plt.savefig(save_path)
+ print(f"图片已保存至:{save_path}")
+ plt.show()
diff --git a/examples/tadf/TADF_angle/config/angle.yaml b/examples/tadf/TADF_angle/config/angle.yaml
new file mode 100644
index 0000000000..8a11d2f4d3
--- /dev/null
+++ b/examples/tadf/TADF_angle/config/angle.yaml
@@ -0,0 +1,60 @@
+defaults:
+ - ppsci_default
+ - TRAIN: train_default
+ - TRAIN/ema: ema_default
+ - TRAIN/swa: swa_default
+ - EVAL: eval_default
+ - INFER: infer_default
+ - hydra/job/config/override_dirname/exclude_keys: exclude_keys_default
+ - _self_
+
+hydra:
+ run:
+ # dynamic output directory according to running time and override name
+ dir: outputs_angle/${now:%Y-%m-%d}/${now:%H-%M-%S}/${hydra.job.override_dirname}
+ job:
+ name: ${mode} # name of logfile
+ chdir: false # keep current working directory unchanged
+ callbacks:
+ init_callback:
+ _target_: ppsci.utils.callbacks.InitCallback
+ sweep:
+ # output directory for multirun
+ dir: ${hydra.run.dir}
+ subdir: ./
+
+# general settings
+mode: eval # running mode: train/eval
+seed: 42
+output_dir: ${hydra:run.dir}
+log_freq: 20
+use_tbd: false
+data_dir: "./angle.dat"
+sim_dir: "./smis.txt"
+
+# model settings
+MODEL:
+ output_keys: ["u"]
+ activation: "relu"
+ dropout: 0.5
+
+# training settings
+TRAIN:
+ epochs: 200
+ iters_per_epoch: 20
+ save_freq: 100
+ eval_during_train: False
+ batch_size: 8
+ learning_rate: 0.01
+ weight_decay: 1e-5
+ pretrained_model_path: null
+ checkpoint_path: null
+ k: 9
+ i: 2
+
+# evaluation settings
+EVAL:
+ test_size: 0.1
+ pretrained_model_path: null
+ batch_size: 8
+ seed: 20
diff --git a/examples/tadf/TADF_f/config/f.yaml b/examples/tadf/TADF_f/config/f.yaml
new file mode 100644
index 0000000000..02b6cf250b
--- /dev/null
+++ b/examples/tadf/TADF_f/config/f.yaml
@@ -0,0 +1,61 @@
+defaults:
+ - ppsci_default
+ - TRAIN: train_default
+ - TRAIN/ema: ema_default
+ - TRAIN/swa: swa_default
+ - EVAL: eval_default
+ - INFER: infer_default
+ - hydra/job/config/override_dirname/exclude_keys: exclude_keys_default
+ - _self_
+
+hydra:
+ run:
+ # dynamic output directory according to running time and override name
+ dir: outputs_f/${now:%Y-%m-%d}/${now:%H-%M-%S}/${hydra.job.override_dirname}
+ job:
+ name: ${mode} # name of logfile
+ chdir: false # keep current working directory unchanged
+ callbacks:
+ init_callback:
+ _target_: ppsci.utils.callbacks.InitCallback
+ sweep:
+ # output directory for multirun
+ dir: ${hydra.run.dir}
+ subdir: ./
+
+# general settings
+mode: eval # running mode: train/eval
+seed: 42
+output_dir: ${hydra:run.dir}
+log_freq: 20
+use_tbd: false
+VIV_DATA_PATH: "./f.dat"
+data_dir: "./f.dat"
+sim_dir: "./smis.txt"
+
+# model settings
+MODEL:
+ output_keys: ["u"]
+ activation: "relu"
+ dropout: 0.5
+
+# training settings
+TRAIN:
+ epochs: 200
+ iters_per_epoch: 20
+ save_freq: 100
+ eval_during_train: False
+ batch_size: 8
+ learning_rate: 0.0001
+ weight_decay: 1e-5
+ pretrained_model_path: null
+ checkpoint_path: null
+ k: 9
+ i: 2
+
+# evaluation settings
+EVAL:
+ test_size: 0.1
+ pretrained_model_path: null
+ batch_size: 8
+ seed: 20
diff --git a/examples/tadf/TADF_f/f.py b/examples/tadf/TADF_f/f.py
new file mode 100644
index 0000000000..5221d079ba
--- /dev/null
+++ b/examples/tadf/TADF_f/f.py
@@ -0,0 +1,22 @@
+import hydra
+from f_model import evaluate
+from f_model import featurize_molecules
+from f_model import load_data
+from f_model import train
+from omegaconf import DictConfig
+
+
+@hydra.main(version_base=None, config_path="./config", config_name="f.yaml")
+def main(cfg: DictConfig):
+ data, smis = load_data(cfg)
+ X = featurize_molecules(smis)
+ if cfg.mode == "train":
+ train(cfg, X, data)
+ elif cfg.mode == "eval":
+ evaluate(cfg, X, data)
+ else:
+ raise ValueError(f"cfg.mode should be 'train'or 'eval', but got '{cfg.mode}'")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/tadf/TADF_f/f_model.py b/examples/tadf/TADF_f/f_model.py
new file mode 100644
index 0000000000..8dec8c576a
--- /dev/null
+++ b/examples/tadf/TADF_f/f_model.py
@@ -0,0 +1,217 @@
+import os
+
+import matplotlib.pyplot as plt
+import numpy as np
+import paddle
+import rdkit.Chem as Chem
+from omegaconf import DictConfig
+from rdkit.Chem import rdFingerprintGenerator
+from sklearn.decomposition import PCA
+from sklearn.model_selection import train_test_split
+
+import ppsci
+
+os.environ["HYDRA_FULL_ERROR"] = "1"
+os.environ["KMP_DUPLICATE_LIB_OK"] = "True"
+plt.rcParams["axes.unicode_minus"] = False
+plt.rcParams["font.sans-serif"] = ["DejaVu Sans"]
+
+# Data preparation
+def load_data(cfg):
+ data_dir = cfg.data_dir
+ sim_dir = cfg.sim_dir
+ f_dat_path = os.path.join(data_dir)
+ smis_txt_path = os.path.join(sim_dir)
+
+ data = []
+ with open(f_dat_path) as f:
+ for line in f:
+ num = float(line.strip())
+ data.append(num)
+ smis = []
+ with open(smis_txt_path) as f:
+ for line in open("./smis.txt"):
+ smis.append(line.strip())
+
+ return data, smis
+
+
+def featurize_molecules(smis):
+ vectors = []
+ del_mol = []
+ for s in smis:
+ mol = Chem.MolFromSmiles(s)
+ try:
+ generator = rdFingerprintGenerator.GetMorganGenerator(radius=2, fpSize=2048)
+ fp = generator.GetFingerprint(mol)
+ _input = np.array(list(map(int, fp.ToBitString())))
+ vectors.append(_input)
+ except Exception as e:
+ print(f"Error processing {s}: {e}")
+ del_mol.append(s)
+ pca = PCA(n_components=0.99)
+ pca.fit(vectors)
+ X = paddle.to_tensor(pca.transform(vectors))
+ return X
+
+
+def train(cfg: DictConfig, X, data):
+ # k-fold cross validation splitter
+ def k_fold(k, i, X, Y):
+ fold_size = tuple(X.shape)[0] // k
+ val_start = i * fold_size
+ if i != k - 1:
+ val_end = (i + 1) * fold_size
+ x_val, y_val = X[val_start:val_end], Y[val_start:val_end]
+ x_train = np.concatenate((X[0:val_start], X[val_end:]), axis=0)
+ y_train = np.concatenate((Y[0:val_start], Y[val_end:]), axis=0)
+ else:
+ x_val, y_val = X[val_start:], Y[val_start:]
+ x_train = X[0:val_start]
+ y_train = Y[0:val_start]
+ return x_train, y_train, x_val, y_val
+
+ Y = paddle.to_tensor(data)
+ x_train, y_train, x_test, y_test = k_fold(cfg.TRAIN.k, cfg.TRAIN.i, X, Y)
+ # Prepare feature dictionary
+ x_train = paddle.to_tensor(x_train, dtype="float32")
+ x = {
+ f"key_{i}": paddle.unsqueeze(
+ paddle.to_tensor(x_train[:, i], dtype="float32"), axis=1
+ )
+ for i in range(x_train.shape[1])
+ }
+ y_train = paddle.unsqueeze(paddle.to_tensor(y_train, dtype="float32"), axis=1)
+
+ # Build supervised constraint
+ sup = ppsci.constraint.SupervisedConstraint(
+ dataloader_cfg={
+ "dataset": {
+ "name": "IterableNamedArrayDataset",
+ "input": x,
+ "label": {"u": y_train},
+ },
+ "batch_size": cfg.TRAIN.batch_size,
+ },
+ loss=ppsci.loss.MSELoss("mean"),
+ name="sup",
+ )
+ # Set model architecture parameters
+ hidden_size = [587, 256]
+ num_layers = None
+
+ # Instantiate TADF model
+ model = ppsci.arch.TADF(
+ input_keys=tuple(x.keys()),
+ hidden_size=hidden_size,
+ num_layers=num_layers,
+ **cfg.MODEL,
+ )
+ optimizer = ppsci.optimizer.optimizer.Adam(
+ cfg.TRAIN.learning_rate,
+ beta1=0.9,
+ beta2=0.99,
+ weight_decay=cfg.TRAIN.weight_decay,
+ )(model)
+
+ # Build solver for training
+ solver = ppsci.solver.Solver(
+ model,
+ constraint={sup.name: sup},
+ optimizer=optimizer,
+ epochs=cfg.TRAIN.epochs,
+ eval_during_train=False,
+ iters_per_epoch=cfg.TRAIN.iters_per_epoch,
+ )
+ try:
+ solver.train()
+ except Exception as ex:
+ print(ex)
+
+
+def evaluate(cfg: DictConfig, X, data):
+ y_full = paddle.to_tensor(data, dtype="float32")
+ X_np = X.numpy()
+ y_np = y_full.numpy()
+ X_train_np, X_test_np, y_train_np, y_test_np = train_test_split(
+ X_np,
+ y_np,
+ test_size=cfg.EVAL.test_size,
+ random_state=cfg.EVAL.seed,
+ )
+ x_test = paddle.to_tensor(X_test_np, dtype="float32")
+ y_test = paddle.to_tensor(y_test_np, dtype="float32")
+
+ x_dict = {
+ f"key_{i}": paddle.unsqueeze(x_test[:, i], axis=1)
+ for i in range(x_test.shape[1])
+ }
+
+ test_validator = ppsci.validate.SupervisedValidator(
+ dataloader_cfg={
+ "dataset": {
+ "name": "IterableNamedArrayDataset",
+ "input": x_dict,
+ "label": {"u": paddle.unsqueeze(y_test, axis=1)},
+ },
+ "batch_size": cfg.EVAL.batch_size,
+ "shuffle": False,
+ },
+ loss=ppsci.loss.MSELoss("mean"),
+ metric={
+ "MAE": ppsci.metric.MAE(),
+ "RMSE": ppsci.metric.RMSE(),
+ "R2": ppsci.metric.R2Score(),
+ },
+ name="test_eval",
+ )
+ validators = {"test_eval": test_validator}
+
+ model = ppsci.arch.TADF(
+ input_keys=tuple(x_dict.keys()),
+ hidden_size=[587, 256],
+ num_layers=None,
+ **cfg.MODEL,
+ )
+
+ solver = ppsci.solver.Solver(
+ model,
+ validator=validators,
+ cfg=cfg,
+ )
+
+ _, metric_dict = solver.eval()
+
+ ypred = model(x_dict)["u"].numpy()
+ ytrue = paddle.unsqueeze(y_test, axis=1).numpy()
+
+ mae = metric_dict["MAE"]["u"]
+ rmse = metric_dict["RMSE"]["u"]
+ r2 = metric_dict["R2"]["u"]
+
+ print("Evaluation metrics:")
+ print(f"MAE: {mae:.4f}")
+ print(f"RMSE: {rmse:.4f}")
+ print(f"R2: {r2:.4f}")
+
+ plt.scatter(
+ ytrue,
+ ypred,
+ s=15,
+ color="royalblue",
+ marker="s",
+ linewidth=1,
+ )
+ plt.plot(
+ [ytrue.min(), ytrue.max()],
+ [ytrue.min(), ytrue.max()],
+ "r-",
+ lw=1,
+ )
+ plt.legend(title=f"R²={r2:.3f}\n\nMAE={mae:.3f}")
+ plt.xlabel("Test θ(°)")
+ plt.ylabel("Predicted θ(°)")
+ save_path = "test_f.png"
+ plt.savefig(save_path)
+ print(f"图片已保存至:{save_path}")
+ plt.show()
diff --git a/examples/tadf/requirements.txt b/examples/tadf/requirements.txt
new file mode 100644
index 0000000000..180e2a38c4
--- /dev/null
+++ b/examples/tadf/requirements.txt
@@ -0,0 +1,2 @@
+rdkit
+scikit-learn
diff --git a/mkdocs.yml b/mkdocs.yml
index 5260fef712..8a81b66783 100644
--- a/mkdocs.yml
+++ b/mkdocs.yml
@@ -122,6 +122,7 @@ nav:
- Moflow: zh/examples/moflow.md
- IFM: zh/examples/ifm.md
- Synthemol: zh/examples/synthemol.md
+ - TADF: zh/examples/tadf.md
- API 文档:
- ppsci:
diff --git a/ppsci/arch/__init__.py b/ppsci/arch/__init__.py
index 5cb698a2a7..9f46d06b4c 100644
--- a/ppsci/arch/__init__.py
+++ b/ppsci/arch/__init__.py
@@ -62,6 +62,7 @@
from ppsci.arch.velocitygan import VelocityGenerator # isort:skip
from ppsci.arch.moflow_net import MoFlowNet, MoFlowProp # isort:skip
from ppsci.utils import logger # isort:skip
+from ppsci.arch.tadf import TADF # isort:skip
from ppsci.arch.regdgcnn import RegDGCNN # isort:skip
from ppsci.arch.regpointnet import RegPointNet # isort:skip
from ppsci.arch.ifm_mlp import IFMMLP # isort:skip
@@ -113,6 +114,7 @@
"RosslerEmbedding",
"SFNONet",
"SPINN",
+ "TADF",
"TFNO1dNet",
"TFNO2dNet",
"TFNO3dNet",
diff --git a/ppsci/arch/tadf.py b/ppsci/arch/tadf.py
new file mode 100644
index 0000000000..4daeb5eb05
--- /dev/null
+++ b/ppsci/arch/tadf.py
@@ -0,0 +1,60 @@
+from typing import Dict
+from typing import Optional
+from typing import Tuple
+from typing import Union
+
+import paddle.nn as nn
+
+from ppsci.arch.mlp import MLP
+
+
+class TADF(MLP):
+ def __init__(
+ self,
+ input_keys: Tuple[str, ...],
+ output_keys: Tuple[str, ...],
+ num_layers: int,
+ hidden_size: Union[int, Tuple[int, ...]],
+ activation: str = "tanh",
+ skip_connection: bool = False,
+ weight_norm: bool = False,
+ input_dim: Optional[int] = None,
+ output_dim: Optional[int] = None,
+ periods: Optional[Dict[str, tuple]] = None,
+ fourier: Optional[Dict[str, Union[float, int]]] = None,
+ random_weight: Optional[Dict[str, float]] = None,
+ dropout: Optional[float] = 0.5,
+ ):
+
+ super().__init__(
+ input_keys=input_keys,
+ output_keys=output_keys,
+ num_layers=num_layers,
+ hidden_size=hidden_size,
+ activation=activation,
+ skip_connection=skip_connection,
+ weight_norm=weight_norm,
+ input_dim=input_dim,
+ output_dim=output_dim,
+ periods=periods,
+ fourier=fourier,
+ random_weight=random_weight,
+ )
+
+ self.dropout = nn.Dropout(p=dropout)
+
+ def forward_tensor(self, x):
+ y = x
+ skip = None
+ for i, linear in enumerate(self.linears):
+ y = linear(y)
+ if self.skip_connection and i % 2 == 0:
+ if skip is not None:
+ skip = y
+ y = y + skip
+ else:
+ skip = y
+ y = self.acts[i](y)
+ y = self.dropout(y)
+ y = self.last_fc(y)
+ return y