This application supports run-time plugins for searching databases.
A demonstration video can be seen here.
- Download the latest package here (1.2.2).
- Open the Garuda Platform, load Gadget Installer
 .
. - Load the package to the using the Gadget Installer
 .
.
- Generated documentation
- Fixed the join table issues
- Added UUID use for setting import settings
How to import data from a file?
RONE accepts two types of file formats, comma-separated value and tab-delimited text.
-
To import import a file, select File > Import > from File
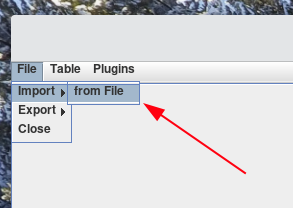
-
Navigate to the file that you are interested in importing. Select the file and then click Open.
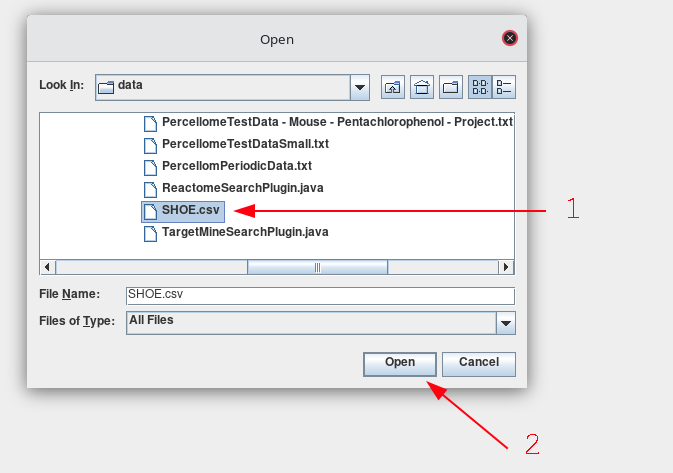
-
Select the format for the file that you are trying to load.
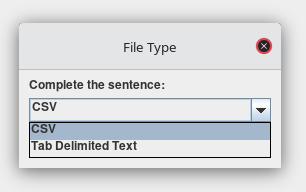
-
Once the file format has been selected, press OK.
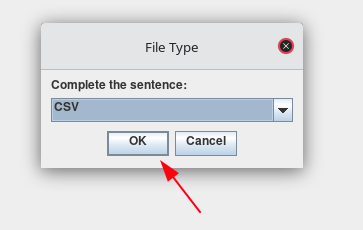
-
Your data will be loaded from the file and presented to you in a new tab.
How to import data from a Garuda Gadget?
Formats that RONE accepts as inputs
| File Type | File Format |
|---|---|
| Genelist | TXT |
| Genelist | CSV |
| Ensemble | TXT |
| Ensemble | CSV |
How to add/remove/edit import settings for other Garuda Gadgets?
-
In your file explorer, navigate to the folder where Garuda Platform is located.
-
Locate and open the folder/file "11136dd6-baa0-49c0-9c1e-d2bec673eec6/config.txt".
You should see the following:
Garuda.GeneMapper.seperator=, Garuda.Reactome\ gadget.seperator=, Garuda.GeneMapper.column_headers=Gene, NM, TF, Region, Strand, MA Score, PSSM Score, ID, Motif, Similarity, Pareto Garuda.Reactome\ gadget.column_headers=Name, Species, Disease Association, Inferred Association, Pathways Garuda.Reactome\ gadget.skip_header=true Garuda.GeneMapper.skip_header=true
-
To add an import setting for a gadget, you will need to add three lines into the configuration file. You can use its UUID (preferable), or you can use the gadget's name to reference the settings for your gadget. In the above case, the gadgets name was used "Reactome gadget". In the following case it will be "MyExampleGadget".
-
Firstly, the "column_headers" refer to the column names of the data provided by the gadget.
Garuda.MyExampleGadget.column_headers=Example Header 1, Example Header 2, Example Header...
-
Secondly, the "seperator" refer the character used to seperate cells. For a CSV-file, that would be ",".
Garuda.MyExampleGadget.seperator=,
-
Finally, the "skip_header" refer tell RONE weather or not the first row of data being imported should be discarded. This is useful if the column headers are contained in the first-row of the incoming data. This value can ONLY be either "true" or "false".
Garuda.MyExampleGadget.skip_header=true
-
Firstly, the "column_headers" refer to the column names of the data provided by the gadget.
How to export data to a file?
RONE only supports exporting data to a comma-separated value file.
-
To export data from a table to a file. Select the cells that you would like to export. If no cells are selected, the whole table will be used at the selection.
-
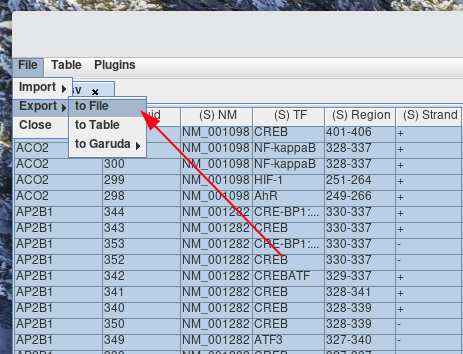
Select File > Export > to File
-
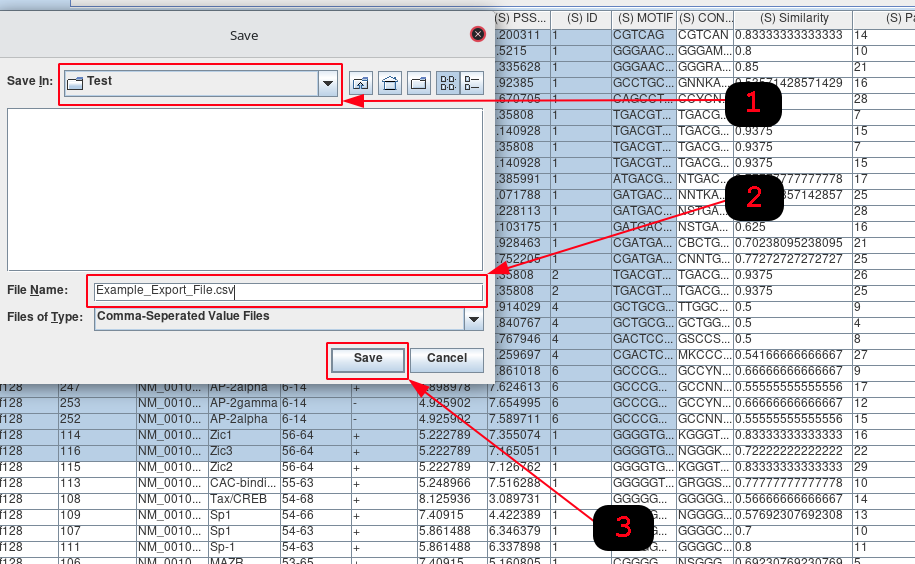
- Navigate to the directory where you would like to store the table/selection.
- Insert the name for the new file that will be generated.
- Click Save.
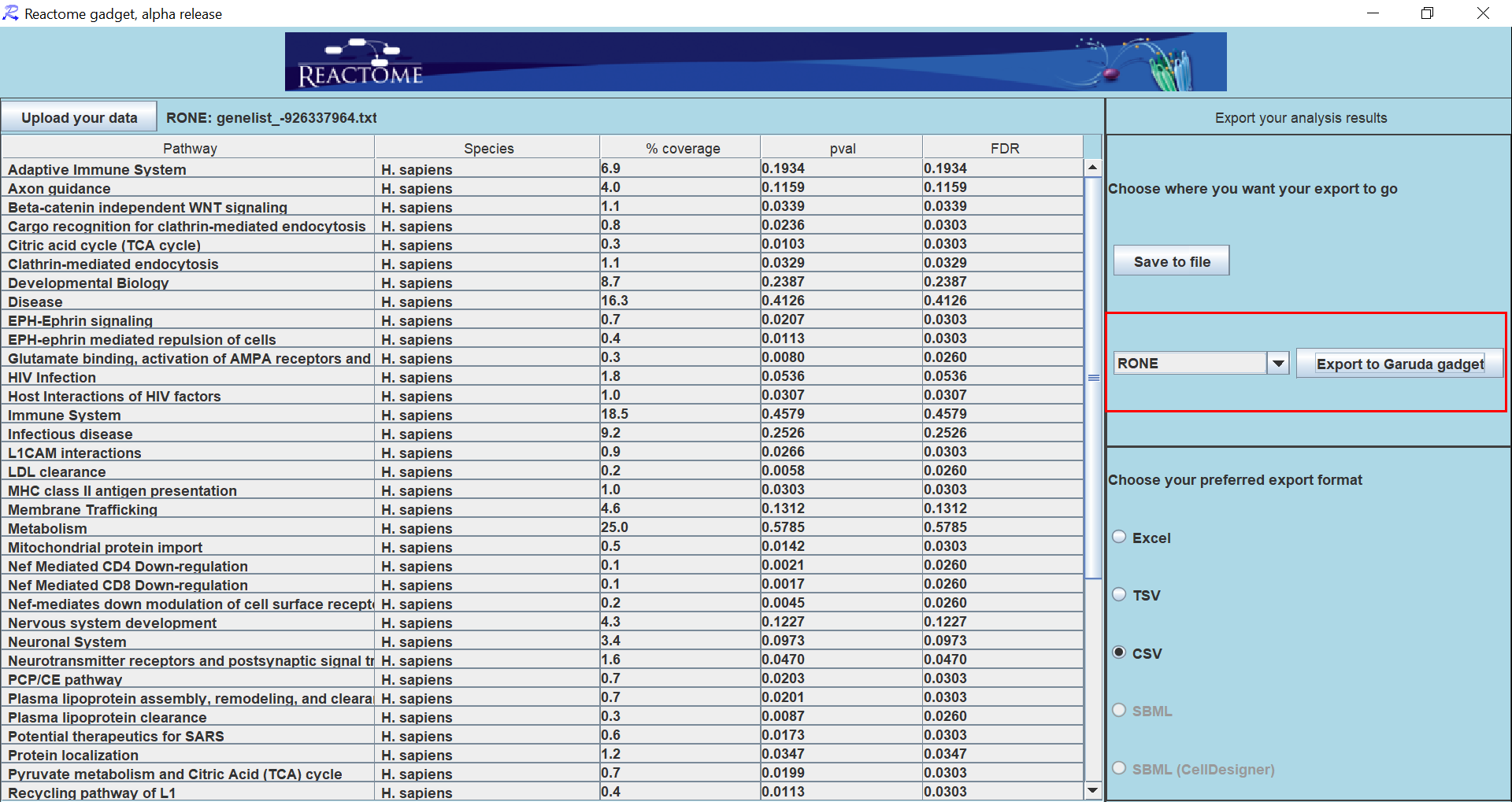
How to export data to a Garuda Gadget?
-
To export data from a table to a gadget. Select the cells that you would like to export. If no cells are selected, the whole table will be as the selection.

-
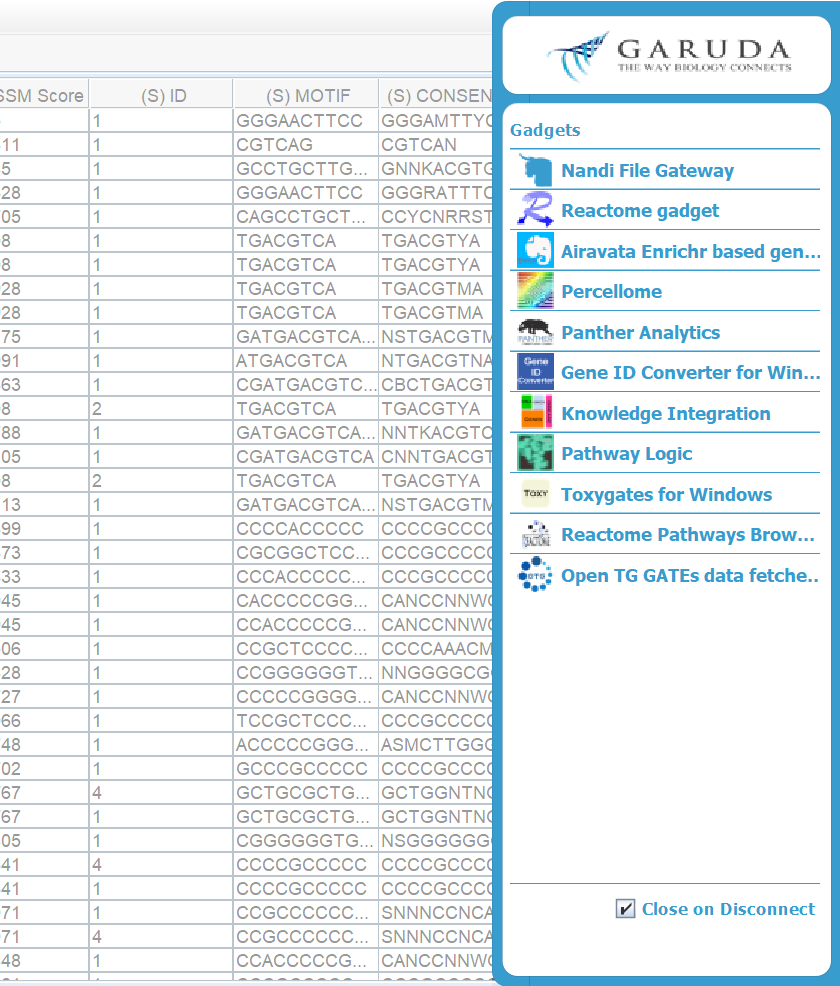
Navigate to File > Export > to Garuda > (export Setting). Select the export option with the desired file type, and file format.
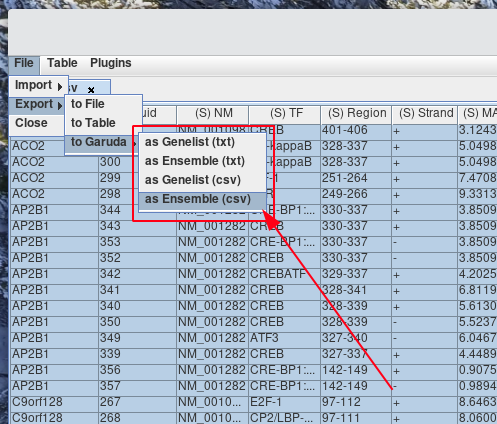
-
To export data from a table to a gadget. Select the cells that you would like to export. If no cells are selected, the whole table will be as the selection.
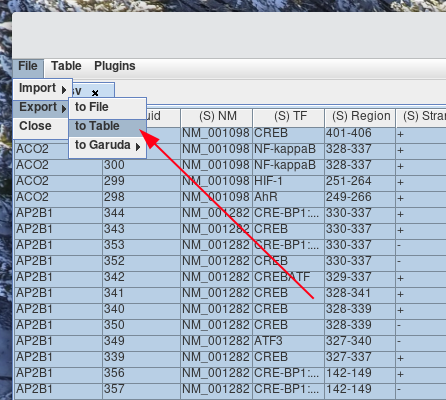
How to export data to a new table?
-
To export data from a table to a file. Select the cells that you would like to export. If no cells are selected, the whole table will be as the selection.

-
To import import a file, select File > Export > to File
-
-
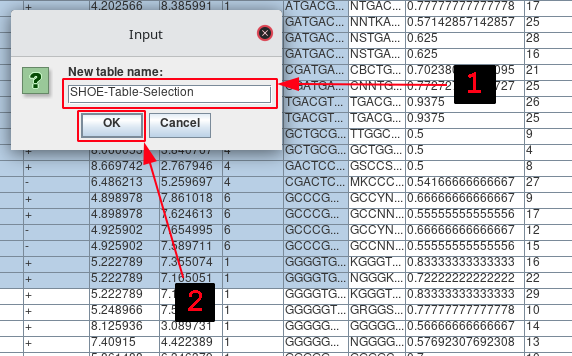
Enter the name for the new table.
-
Press OK.
-
Enter the name for the new table.
-
The selected data will be presented in the new tab.
How to sort a table?
Tables can be sorted by their columns in ascending and descending order. Further, order by operations can be employed as well; sorting by column A, then by column B.
-
Right-click the column header for the column that you would like to sort.
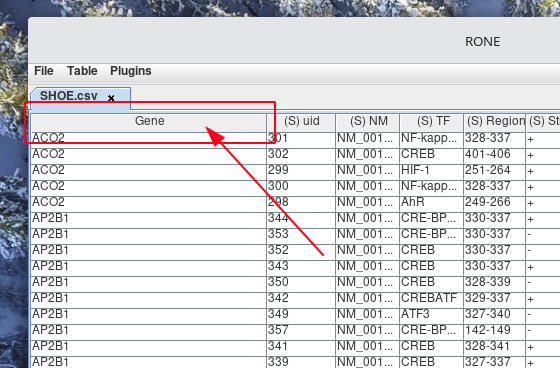
-
Left-click the name of the Sort by column header from the pop-up menu that appears. By default, the column will sort in ascending order.
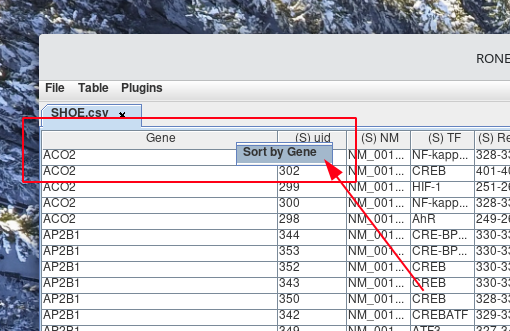
-
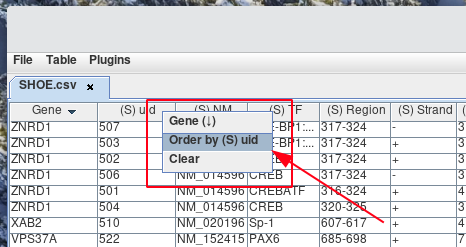
If you would like to sort the column in descending order. Then Right-click the column header from step 1. Next, Left-click the name of the column header from the pop-up menu that appears. You will notice next to the column header, either (↑) or (↓) denotes if the column is currently sorted in ascending or descending order.
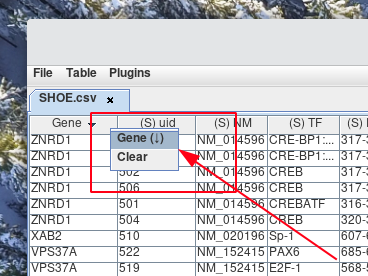
-
After sorting by column A, you can sort column B, then C, D, and so on. This is accomplished by Right-clicking the column you next want to sort by. Then Left-click the name of the Order by column header from the pop-up menu that appears. You will notice next to the column header, either (↑) or (↓) denotes if the column is currently being sorted in ascending or descending order.
-
If you no longer wish to have the rows ordered with the current sort by, order by filter, you can clear it. This is accomplished by Right-clicking a column header and selecting Clear from the pop-up menu.
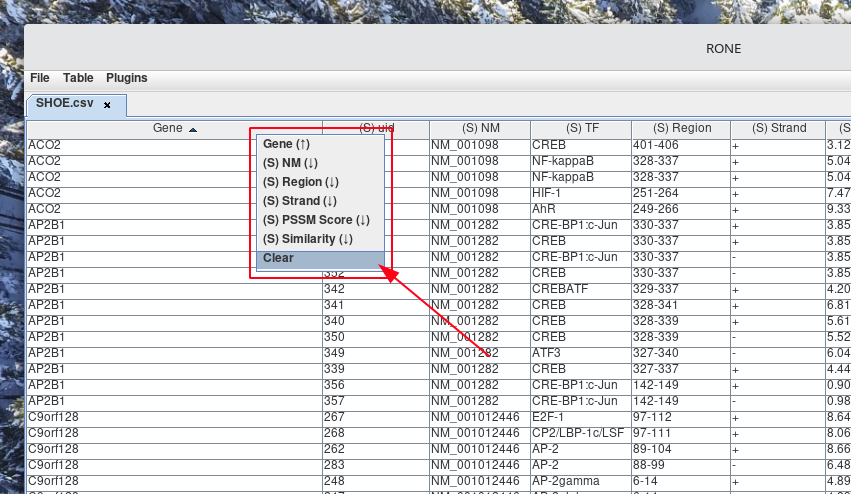
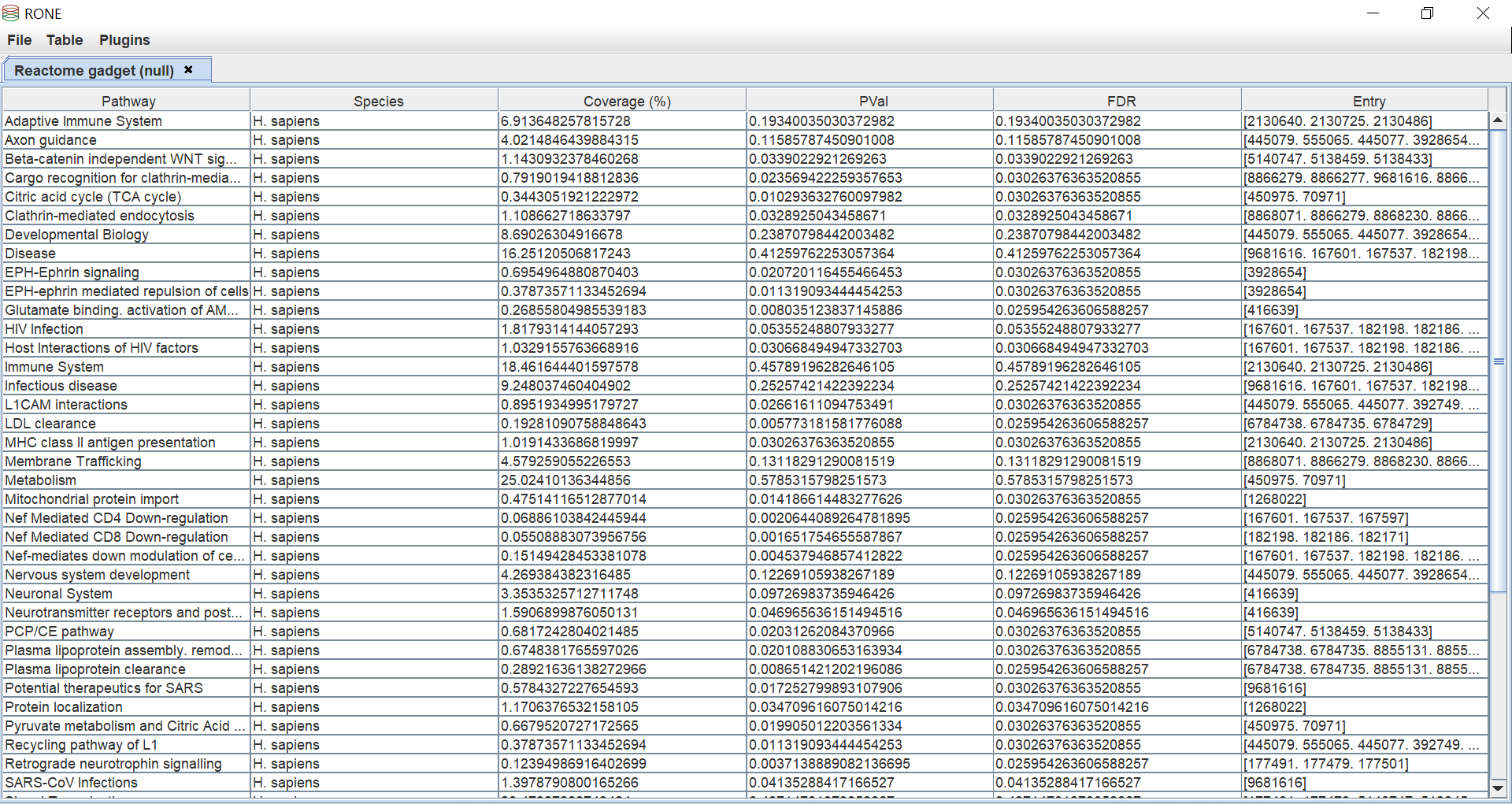
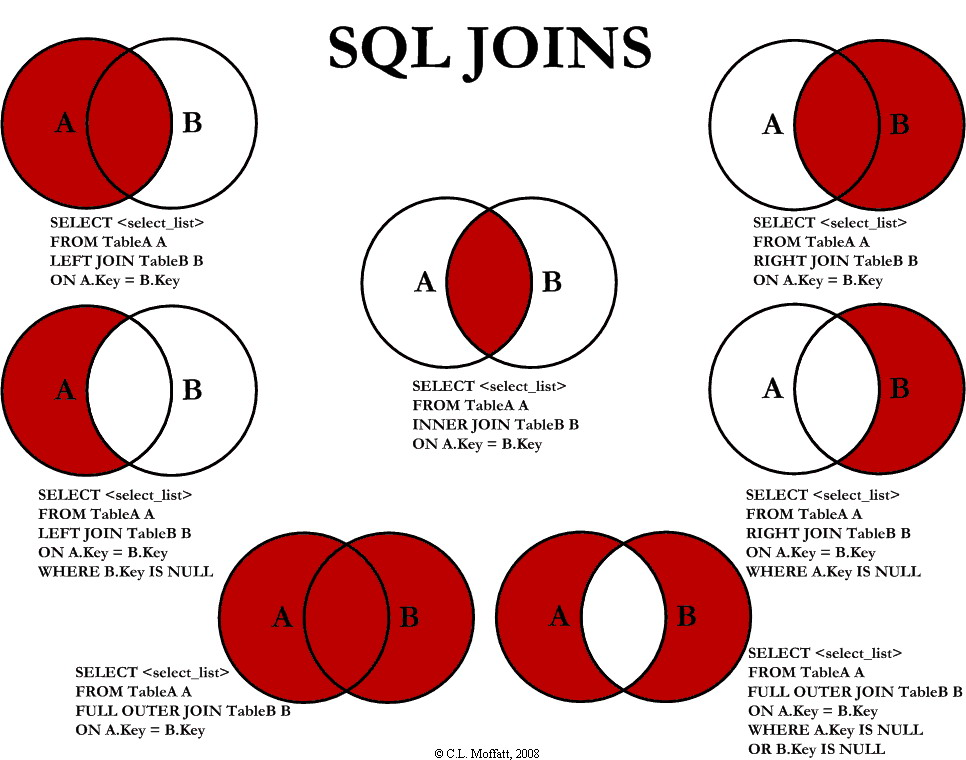
How to join tables together?
RONE support the standard join operations offered by SQL.
- Left Inclusive
- Left Exclusive
- Right Inclusive
- Right Exclusive
- Full Outer Inclusion
- Full Outer Exclusion
- Inner
Note, RONE only supports "=" constraints.
SELECT column name(s)
FROM table1
LEFT JOIN table2
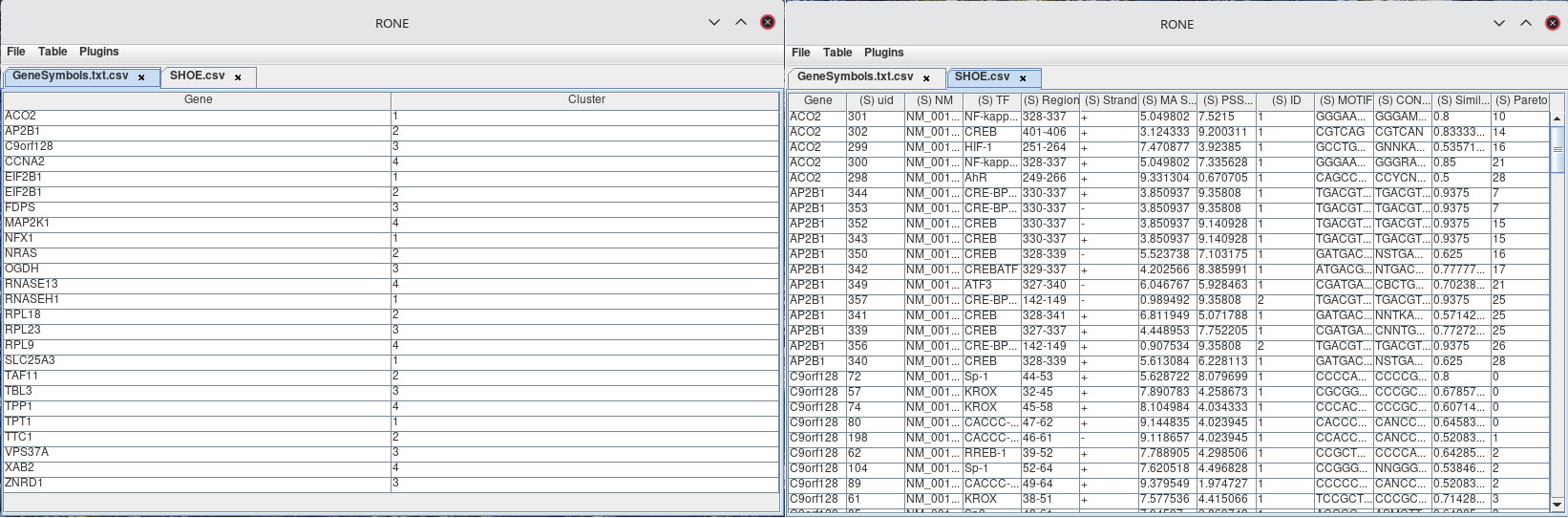
ON table1.column_name = table2.column_name;To be able to join two tables together, you will need at least two tables. The tables will ideally contain two columns of the same values. For instance, bellow you see in the SHOE.csv table, there is a column called Gene, and in the table GeneSymbols.txt.csv, there is a column called Gene. The joint will be performed on the same column.
-
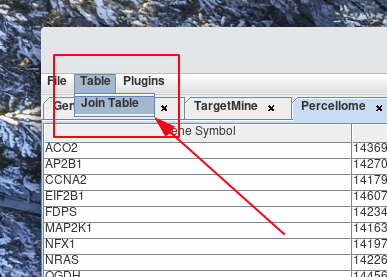
To join two tables together, select Table > Join Table.
-
A new window called Join Table will appear. Using the combo-boxes Table A, and Table B in Table Select section, select the tables you would like to join.
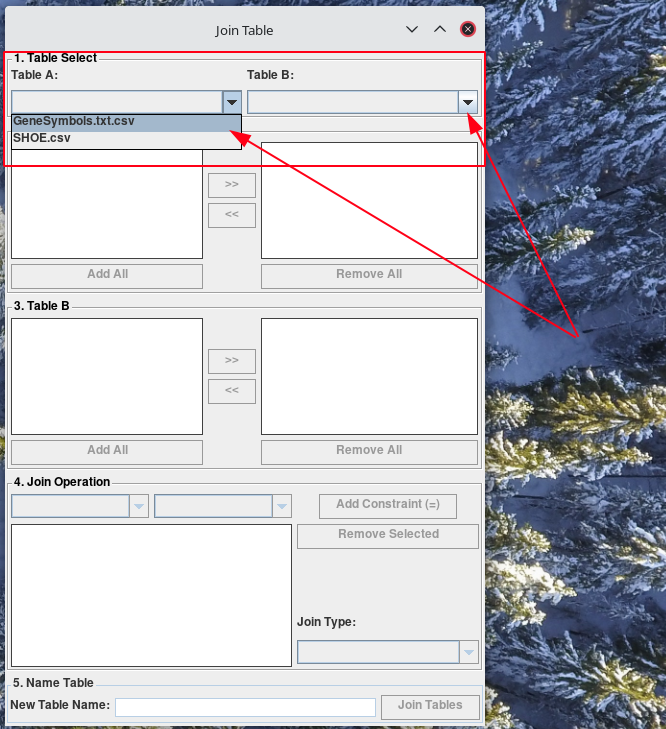
-
Now, you will see that it added the columns of the selected tables into the list on the left in Table A section, and Table B section. The list boxes on the right of Table A section, and Table B section denote columns included to the resulting tab. Each section is an exclude and include list (left and right). You can move columns between the include and exclude columns using the buttons: >>, <<, Add All, Remove All.
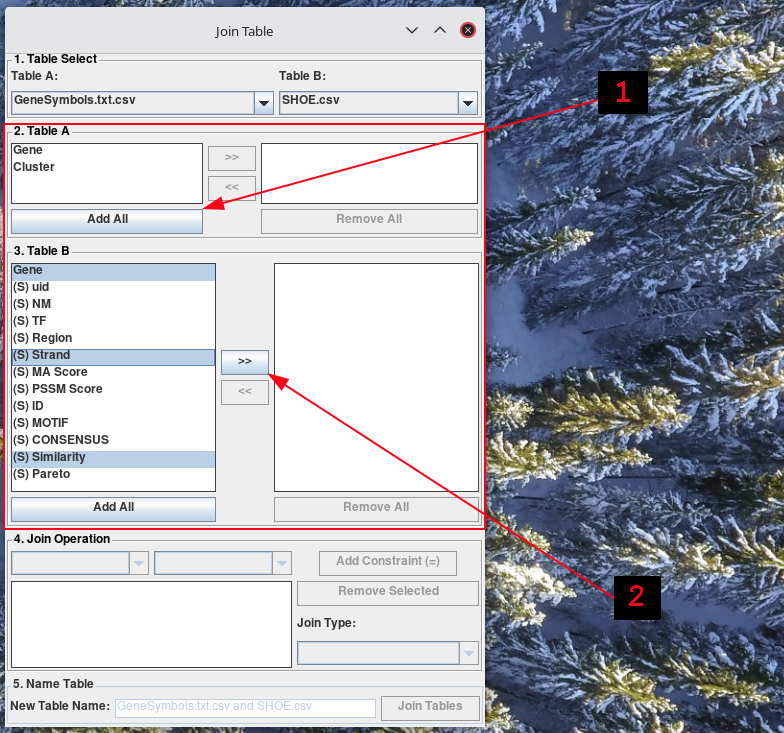
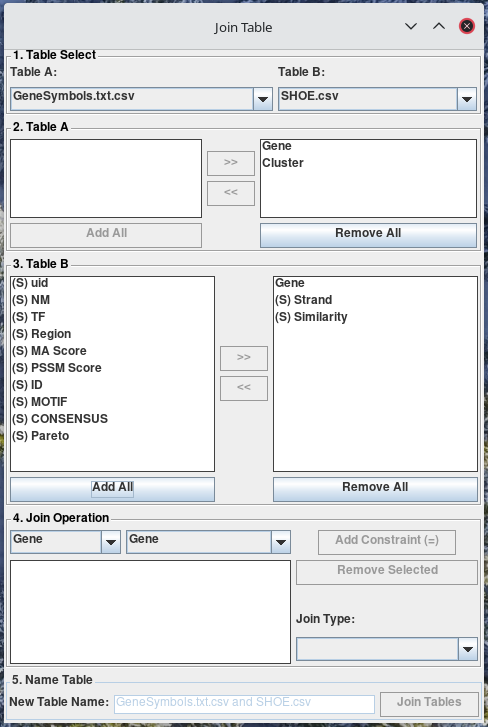
-
From the two combo-boxes at the top of the Join Operation section, select the columns from the tables you would like to constrain. Then, from the combo-box in the bottom-right corner of the Join Operation section, select the type of joint you would like to perform.
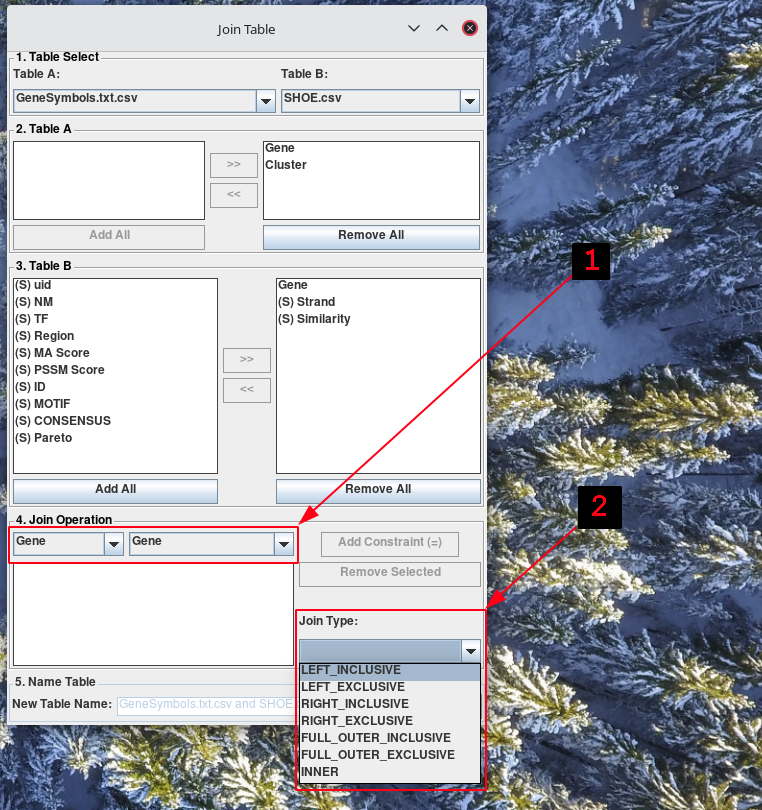
-
-
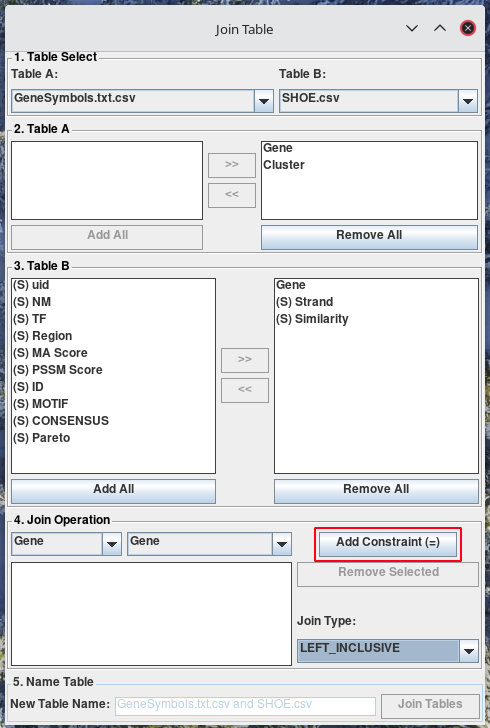
Click Add Constraint (=). You will see the constraint appear in the table. You can add several constraints by simply changing the selected columns, and then clicking Add Constraint (=) again.
-
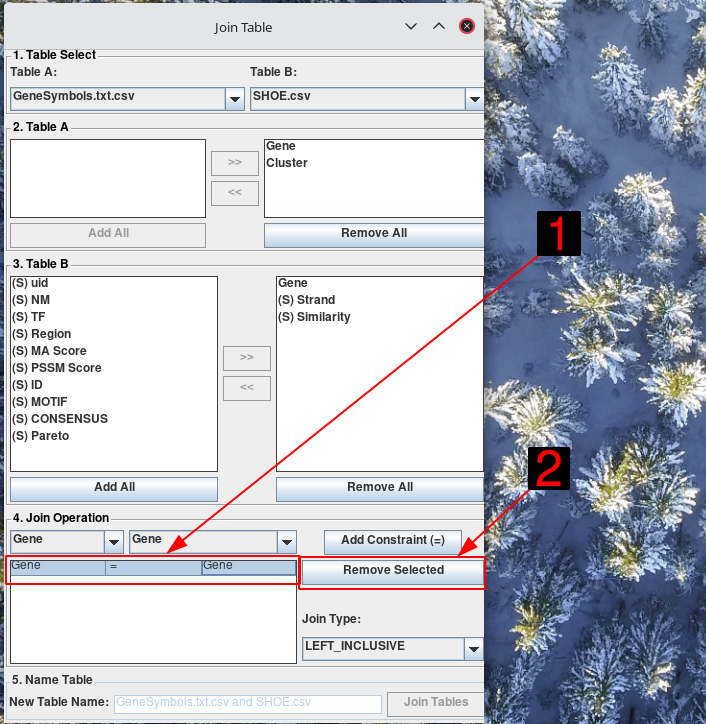
To remove constraints, select the constraint from the table in the Join Operations section. Click Remove Selected
-
Click Add Constraint (=). You will see the constraint appear in the table. You can add several constraints by simply changing the selected columns, and then clicking Add Constraint (=) again.
-
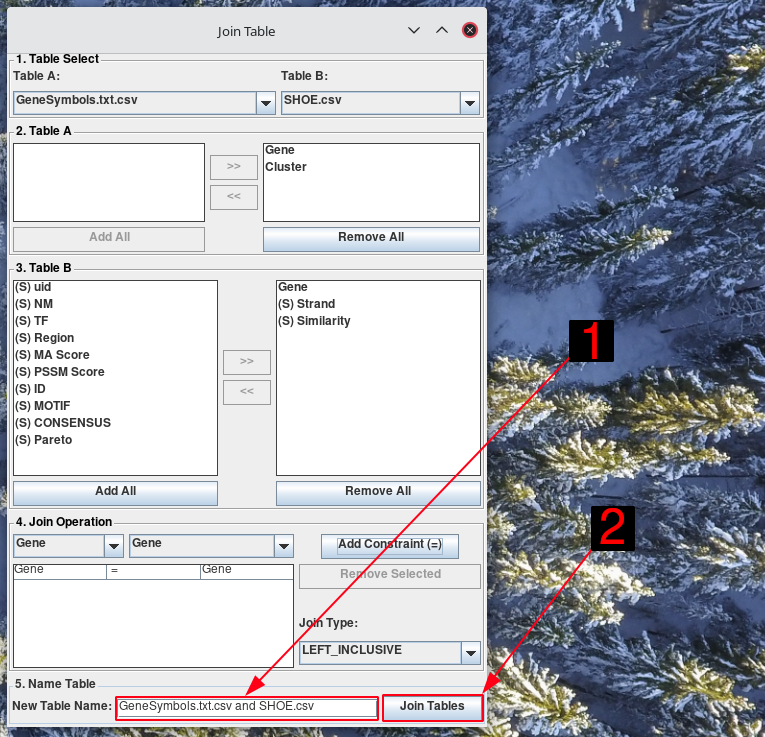
Finally, in the Name Table section, enter the name for the new table. Click Join Table.
Equivelent to
SELECT GeneSymbols.Gene, GeneSymbols.Cluster, SHOE.Gene, SHOE.Strand, SHOE.Similarity FROM GeneSymbols LEFT JOIN SHOE ON GeneSymbols.Gene = SHOE.Gene;
-
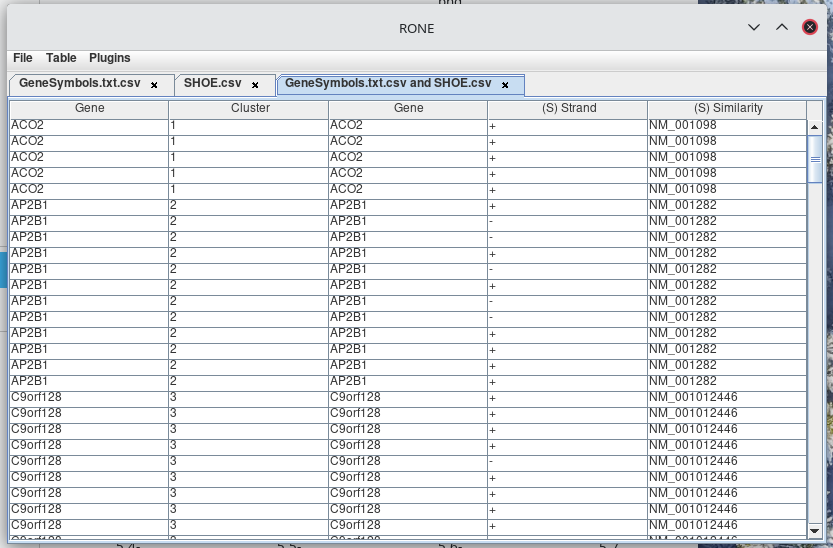
The resulting table can be seen the new tab.
How to use plugins?
When RONE begins, it trys to activate its plugins. If the plugin has been loaded successfully, it can be accessed from the plugin menu.
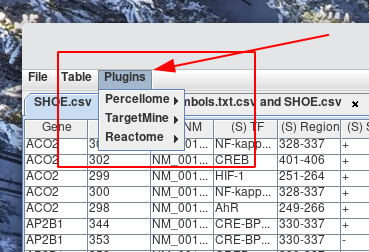

Default plugins
TargetMine
| Input | Output |
|---|---|
| Gene Symbol | Gene Symbol, Primary Identifer, Name, Pathway Identifer, Pathway Name, Organism Name, Gene Pathway Label 1, Gene Pathway Label 2 |
Reactome
| Input | Output |
|---|---|
| Gene Symbols | Gene Symbol, Species Name, Pathway Name, Pathway stId, Pathway Disease Association, Pathway III, Entities Ratio, Entities pValue, Entities FDR |
Percellome
| Input | Output |
|---|---|
| Gene Symbol, Species | Gene Symbol, Probe (Affy ID), Found Gene Symbol |
| Probe IDs, Species | Probe ID (Affy ID), Gene Symbol, Biological Function, Cellular Function, Molecular Function |