diff --git a/docs/no_toc/01-lesson1.md b/docs/no_toc/01-lesson1.md
new file mode 100644
index 0000000..b557a40
--- /dev/null
+++ b/docs/no_toc/01-lesson1.md
@@ -0,0 +1,361 @@
+# Intro to Computing
+
+*Slides that go with this lesson can be found [here](slides/lesson1_slides.html).*
+
+## Goals of the course
+
+- Fundamental concepts in high-level programming languages (R, Python, Julia, WDL, etc.) that is transferable: *How do programs run, and how do we solve problems using functions and data structures?*
+
+- Beginning of data science fundamentals: *How do you translate your scientific question to a data wrangling problem and answer it?*
+
+ 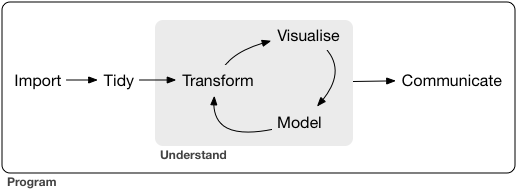{width="450"}
+
+- Find a nice balance between the two throughout the course: we will try to reproduce a figure from a scientific publication using new data.
+
+## What is a computer program?
+
+- A sequence of instructions to manipulate data for the computer to execute.
+
+- A series of translations: English \<-\> Programming Code for Interpreter \<-\> Machine Code for Central Processing Unit (CPU)
+
+We will focus on English \<-\> Programming Code for R Interpreter in this class.
+
+More importantly: **How we organize ideas \<-\> Instructing a computer to do something**.
+
+## A programming language has following elements: {#a-programming-language-has-following-elements}
+
+- Grammar structure (simple building blocks)
+
+- Means of combination to analyze and create content (examples around genomics provided, and your scientific creativity is strongly encouraged!)
+
+- Means of abstraction for modular and reusable content (data structures, functions)
+
+- Culture (emphasis on open-source, collaborative, reproducible code)
+
+Requires a lot of practice to be fluent!
+
+## What is R and why should I use it?
+
+It is a:
+
+- Dynamic programming interpreter
+
+- Highly used for data science, visualization, statistics, bioinformatics
+
+- Open-source and free; easy to create and distribute your content; quirky culture
+
+## R vs. Python as a first language
+
+In terms of our goals, recall:
+
+- Fundamental concepts in high-level programming languages
+
+- Beginning of data science fundamentals
+
+There are a lot of nuances and debates, but I argue that Python is a better learning environment for the former and R is better for the latter.
+
+Ultimately, either should be okay! Perhaps more importantly, *consider what your research group and collaborator are more comfortable with*.
+
+## Posit Cloud Setup
+
+Posit Cloud/RStudio is an Integrated Development Environment (IDE). Think about it as Microsoft Word to a plain text editor. It provides extra bells and whistles to using R that is easier for the user.
+
+Today, we will pay close attention to:
+
+- Script editor: where sequence of instructions are typed and saved as a text document as a R program. To run the program, the console will execute every single line of code in the document.
+
+- Console (interpreter): Instead of giving a entire program in a text file, you could interact with the R Console line by line. You give it one line of instruction, and the console executes that single line. It is what R looks like without RStudio.
+
+- Environment: Often, code will store information *in memory*, and it is shown in the environment. More on this later.
+
+## Using Quarto for your work
+
+Why should we use Quarto for data science work?
+
+- Encourages reproducible workflows
+
+- Code, output from code, and prose combined together
+
+- Extendability to Python, Julia, and more.
+
+More options and guides can be found in [Introduction to Quarto](https://quarto.org/docs/get-started/hello/rstudio.html) .
+
+## Grammar Structure 1: Evaluation of Expressions
+
+- **Expressions** are be built out of **operations** or **functions**.
+
+- Operations and functions combine **data types** to return another data type.
+
+- We can combine multiple expressions together to form more complex expressions: an expression can have other expressions nested inside it.
+
+For instance, consider the following expressions entered to the R Console:
+
+
+```r
+18 + 21
+```
+
+```
+## [1] 39
+```
+
+```r
+max(18, 21)
+```
+
+```
+## [1] 21
+```
+
+```r
+max(18 + 21, 65)
+```
+
+```
+## [1] 65
+```
+
+```r
+18 + (21 + 65)
+```
+
+```
+## [1] 104
+```
+
+```r
+length("ATCG")
+```
+
+```
+## [1] 1
+```
+
+Here, our input **data types** to the operation are **numeric** in lines 1-4 and our input data type to the function is **character** in line 5.
+
+Operations are just functions in hiding. We could have written:
+
+
+```r
+sum(18, 21)
+```
+
+```
+## [1] 39
+```
+
+```r
+sum(18, sum(21, 65))
+```
+
+```
+## [1] 104
+```
+
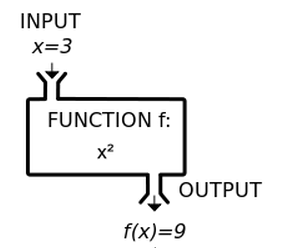
+Remember the function machine from algebra class? We will use this schema to think about expressions.
+
+
+
+If an expression is made out of multiple, nested operations, what is the proper way of the R Console interpreting it? Being able to read nested operations and nested functions as a programmer is very important.
+
+
+```r
+3 * 4 + 2
+```
+
+```
+## [1] 14
+```
+
+```r
+3 * (4 + 2)
+```
+
+```
+## [1] 18
+```
+
+Lastly, a note on the use of functions: a programmer should not need to know how the function is implemented in order to use it - this emphasizes [abstraction and modular thinking](#a-programming-language-has-following-elements), a foundation in any programming language.
+
+## Grammar Structure 2: Storing data types in the global environment
+
+To build up a computer program, we need to store our returned data type from our expression somewhere for downstream use. We can assign a variable to it as follows:
+
+
+```r
+x = 18 + 21
+```
+
+If you enter this in the Console, you will see that in the Environment, the variable `x` has a value of `39`.
+
+### Execution rule for variable assignment
+
+> Evaluate the expression to the right of `=`.
+>
+> Bind variable to the left of `=` to the resulting value.
+>
+> The variable is stored in the environment.
+>
+> `<-` is okay too!
+
+The environment is where all the variables are stored, and can be used for an expression anytime once it is defined. Only one unique variable name can be defined.
+
+The variable is stored in the working memory of your computer, Random Access Memory (RAM). This is temporary memory storage on the computer that can be accessed quickly. Typically a personal computer has 8, 16, 32 Gigabytes of RAM. When we work with large datasets, if you assign a variable to a data type larger than the available RAM, it will not work. More on this later.
+
+Look, now `x` can be reused downstream:
+
+
+```r
+x - 2
+```
+
+```
+## [1] 37
+```
+
+```r
+y = x * 2
+```
+
+## Grammar Structure 3: Evaluation of Functions
+
+A function has a **function name**, **arguments**, and **returns** a data type.
+
+### Execution rule for functions:
+
+> Evaluate the function by its arguments, and if the arguments are functions or contains operations, evaluate those functions or operations first.
+>
+> The output of functions is called the **returned value**.
+
+
+```r
+sqrt(nchar("hello"))
+```
+
+```
+## [1] 2.236068
+```
+
+```r
+(nchar("hello") + 4) * 2
+```
+
+```
+## [1] 18
+```
+
+## Functions to read in data
+
+We are going to read in a Comma Separated Value (CSV) spreadsheet, that contains information about cancer cell lines.
+
+The first line calls the function `read.csv()` with a string argument representing the file path to the CSV file (we are using an URL online, but this is typically done locally), and the returned data type is stored in `metadata` variable. The resulting `metadata` variable is a new data type you have never seen before. It is a **data structure** called a **data frame** that we will be exploring next week. It holds a table of several data types that we can explore.
+
+We run a few functions on `metadata`.
+
+
+```r
+metadata = read.csv("https://github.com/caalo/Intro_to_R/raw/main/classroom_data/CCLE_metadata.csv")
+head(metadata)
+```
+
+```
+## ModelID PatientID CellLineName StrippedCellLineName Age SourceType
+## 1 ACH-000001 PT-gj46wT NIH:OVCAR-3 NIHOVCAR3 60 Commercial
+## 2 ACH-000002 PT-5qa3uk HL-60 HL60 36 Commercial
+## 3 ACH-000003 PT-puKIyc CACO2 CACO2 72 Commercial
+## 4 ACH-000004 PT-q4K2cp HEL HEL 30 Commercial
+## 5 ACH-000005 PT-q4K2cp HEL 92.1.7 HEL9217 30 Commercial
+## 6 ACH-000006 PT-ej13Dz MONO-MAC-6 MONOMAC6 64 Commercial
+## SangerModelID RRID DepmapModelType AgeCategory GrowthPattern
+## 1 SIDM00105 CVCL_0465 HGSOC Adult Adherent
+## 2 SIDM00829 CVCL_0002 AML Adult Suspension
+## 3 SIDM00891 CVCL_0025 COAD Adult Adherent
+## 4 SIDM00594 CVCL_0001 AML Adult Suspension
+## 5 SIDM00593 CVCL_2481 AML Adult Mixed
+## 6 SIDM01023 CVCL_1426 AMOL Adult Suspension
+## LegacyMolecularSubtype PrimaryOrMetastasis SampleCollectionSite
+## 1 Metastatic ascites
+## 2 Primary haematopoietic_and_lymphoid_tissue
+## 3 Primary Colon
+## 4 Primary haematopoietic_and_lymphoid_tissue
+## 5 bone_marrow
+## 6 Primary haematopoietic_and_lymphoid_tissue
+## Sex SourceDetail LegacySubSubtype CatalogNumber
+## 1 Female ATCC high_grade_serous HTB-71
+## 2 Female ATCC M3 CCL-240
+## 3 Male ATCC HTB-37
+## 4 Male DSMZ M6 ACC 11
+## 5 Male ATCC M6 HEL9217
+## 6 Male DSMZ M5 ACC 124
+## CCLEName COSMICID PublicComments
+## 1 NIHOVCAR3_OVARY 905933
+## 2 HL60_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 905938
+## 3 CACO2_LARGE_INTESTINE NA
+## 4 HEL_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 907053
+## 5 HEL9217_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE NA
+## 6 MONOMAC6_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 908148
+## WTSIMasterCellID EngineeredModel TreatmentStatus OnboardedMedia PlateCoating
+## 1 2201 MF-001-041 None
+## 2 55 MF-005-001 None
+## 3 NA Unknown MF-015-009 None
+## 4 783 Post-treatment MF-001-001 None
+## 5 NA MF-001-001 None
+## 6 2167 MF-001-001 None
+## OncotreeCode OncotreeSubtype OncotreePrimaryDisease
+## 1 HGSOC High-Grade Serous Ovarian Cancer Ovarian Epithelial Tumor
+## 2 AML Acute Myeloid Leukemia Acute Myeloid Leukemia
+## 3 COAD Colon Adenocarcinoma Colorectal Adenocarcinoma
+## 4 AML Acute Myeloid Leukemia Acute Myeloid Leukemia
+## 5 AML Acute Myeloid Leukemia Acute Myeloid Leukemia
+## 6 AMOL Acute Monoblastic/Monocytic Leukemia Acute Myeloid Leukemia
+## OncotreeLineage
+## 1 Ovary/Fallopian Tube
+## 2 Myeloid
+## 3 Bowel
+## 4 Myeloid
+## 5 Myeloid
+## 6 Myeloid
+```
+
+```r
+nrow(metadata)
+```
+
+```
+## [1] 1864
+```
+
+```r
+ncol(metadata)
+```
+
+```
+## [1] 30
+```
+
+If you don't know what a function does, ask for help:
+
+
+```r
+?nrow
+```
+
+## Tips on Exercises / Debugging
+
+Common errors:
+
+- Syntax error.
+
+- Changing a variable without realizing you did so.
+
+- The function or operation does not accept the input data type.
+
+- It did something else than I expected!
+
+Solutions:
+
+- Where is the problem?
+
+- What kind of problem is it?
+
+- Explain your problem to someone!
diff --git a/docs/no_toc/404.html b/docs/no_toc/404.html
index 1f2e9cb..6c844b7 100644
--- a/docs/no_toc/404.html
+++ b/docs/no_toc/404.html
@@ -4,11 +4,11 @@
- Page not found | Introduction-to-R,-Season-1
-
+ Page not found | Season 1 Introduction to R
+
-
+
@@ -16,7 +16,7 @@
-
+
@@ -28,7 +28,7 @@
-
+
@@ -134,10 +134,10 @@
-
diff --git a/docs/no_toc/About.md b/docs/no_toc/About.md
index 7676a36..67d82a8 100644
--- a/docs/no_toc/About.md
+++ b/docs/no_toc/About.md
@@ -1,43 +1,41 @@
-
-# About the Authors {-}
+# About the Authors {.unnumbered}
These credits are based on our [course contributors table guidelines](https://www.ottrproject.org/more_features.html#giving-credits-to-contributors).
-
-
+
-|Credits|Names|
-|-------|-----|
-|**Pedagogy**||
-|Lead Content Instructor(s)|[FirstName LastName]|
-|Lecturer(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved| Delivered the course in some way - video or audio|
-|Content Author(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved | If any other authors besides lead instructor|
-|Content Contributor(s) (include section name/link in parentheses) - make new line if more than one section involved| Wrote less than a chapter|
-|Content Editor(s)/Reviewer(s) | Checked your content|
-|Content Director(s) | Helped guide the content direction|
-|Content Consultants (include chapter name/link in parentheses or word "General") - make new line if more than one chapter involved | Gave high level advice on content|
-|Acknowledgments| Gave small assistance to content but not to the level of consulting |
-|**Production**||
-|Content Publisher(s)| Helped with publishing platform|
-|Content Publishing Reviewer(s)| Reviewed overall content and aesthetics on publishing platform|
-|**Technical**||
-|Course Publishing Engineer(s)| Helped with the code for the technical aspects related to the specific course generation|
-|Template Publishing Engineers|[Candace Savonen], [Carrie Wright], [Ava Hoffman]|
-|Publishing Maintenance Engineer|[Candace Savonen]|
-|Technical Publishing Stylists|[Carrie Wright], [Ava Hoffman], [Candace Savonen]|
-|Package Developers ([ottrpal]) [Candace Savonen], [John Muschelli], [Carrie Wright]|
-|**Art and Design**||
-|Illustrator(s)| Created graphics for the course|
-|Figure Artist(s)| Created figures/plots for course|
-|Videographer(s)| Filmed videos|
-|Videography Editor(s)| Edited film|
-|Audiographer(s)| Recorded audio|
-|Audiography Editor(s)| Edited audio recordings|
-|**Funding**||
-|Funder(s)| Institution/individual who funded course including grant number|
-|Funding Staff| Staff members who help with funding|
+| Credits | Names |
+|------------------------------------------|------------------------------|
+| **Pedagogy** | |
+| Lead Content Instructor(s) | Chris Lo |
+| Lecturer | Chris Lo |
+| Content Author(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved | If any other authors besides lead instructor |
+| Content Contributor(s) (include section name/link in parentheses) - make new line if more than one section involved | Wrote less than a chapter |
+| Content Editor(s)/Reviewer(s) | Checked your content |
+| Content Director(s) | Helped guide the content direction |
+| Content Consultants (include chapter name/link in parentheses or word "General") - make new line if more than one chapter involved | Gave high level advice on content |
+| Acknowledgments | Gave small assistance to content but not to the level of consulting |
+| **Production** | |
+| Content Publisher(s) | Helped with publishing platform |
+| Content Publishing Reviewer(s) | Reviewed overall content and aesthetics on publishing platform |
+| **Technical** | |
+| Course Publishing Engineer(s) | Helped with the code for the technical aspects related to the specific course generation |
+| Template Publishing Engineers | [Candace Savonen](https://www.cansavvy.com/), [Carrie Wright](https://carriewright11.github.io/), [Ava Hoffman](https://www.avahoffman.com/) |
+| Publishing Maintenance Engineer | [Candace Savonen](https://www.cansavvy.com/) |
+| Technical Publishing Stylists | [Carrie Wright](https://carriewright11.github.io/), [Ava Hoffman](https://www.avahoffman.com/), [Candace Savonen](https://www.cansavvy.com/) |
+| Package Developers ([ottrpal](https://github.com/jhudsl/ottrpal)) [Candace Savonen](https://www.cansavvy.com/), [John Muschelli](https://johnmuschelli.com/), [Carrie Wright](https://carriewright11.github.io/) | |
+| **Art and Design** | |
+| Illustrator(s) | Created graphics for the course |
+| Figure Artist(s) | Created figures/plots for course |
+| Videographer(s) | Filmed videos |
+| Videography Editor(s) | Edited film |
+| Audiographer(s) | Recorded audio |
+| Audiography Editor(s) | Edited audio recordings |
+| **Funding** | |
+| Funder(s) | Institution/individual who funded course including grant number |
+| Funding Staff | Staff members who help with funding |
-
+
```
@@ -112,16 +110,9 @@ These credits are based on our [course contributors table guidelines](https://ww
-[FirstName LastName]: link to personal website
-[John Muschelli]: https://johnmuschelli.com/
-[Candace Savonen]: https://www.cansavvy.com/
-[Carrie Wright]: https://carriewright11.github.io/
-[Ava Hoffman]: https://www.avahoffman.com/
-
-[ottrpal]: https://github.com/jhudsl/ottrpal
-
+```{=html}
+```
diff --git a/docs/no_toc/about-the-authors.html b/docs/no_toc/about-the-authors.html
index 7d97c08..54357e5 100644
--- a/docs/no_toc/about-the-authors.html
+++ b/docs/no_toc/about-the-authors.html
@@ -4,11 +4,11 @@
- About the Authors | Introduction-to-R,-Season-1
-
+ About the Authors | Season 1 Introduction to R
+
-
+
@@ -16,7 +16,7 @@
-
+
@@ -28,7 +28,7 @@
-
+
@@ -134,10 +134,10 @@
-
The course is intended for researchers who want to learn coding for the first time with a data science application, or have explored programming and want to focus on fundamentals.
+
+
Season 1 Introduction to R
+
September, 2023
-
-
1.2 Curriculum
-
The course covers fundamentals of R, a high-level programming language, and use it to wrangle data for analysis and visualization. The programming skills you will learn are transferable to learn more about R independently and other high-level languages such as Python. At the end of the class, you will be reproducing analysis from a scientific publication!
+
+
Chapter 1 About this Course
+
+
1.1 Curriculum
+
The course covers fundamentals of R, a high-level programming language, and use it to wrangle data for analysis and visualization.
+
+
+
1.2 Target Audience
+
The course is intended for researchers who want to learn coding for the first time with a data science application, or have explored programming and want to focus on fundamentals.
diff --git a/docs/no_toc/index.md b/docs/no_toc/index.md
index 6a8d644..4aa0dd0 100644
--- a/docs/no_toc/index.md
+++ b/docs/no_toc/index.md
@@ -1,26 +1,25 @@
---
-title: "Course Name"
+title: "Season 1 Introduction to R"
date: "September, 2023"
site: bookdown::bookdown_site
documentclass: book
bibliography: [book.bib]
biblio-style: apalike
link-citations: yes
-description: "Description about Course/Book."
+description: ""
favicon: assets/dasl_favicon.ico
output:
bookdown::word_document2:
toc: true
---
-# About this Course {-}
+# About this Course
+## Curriculum
-## Available course formats
+The course covers fundamentals of R, a high-level programming language, and use it to wrangle data for analysis and visualization.
-This course is available in multiple formats which allows you to take it in the way that best suites your needs. You can take it for certificate which can be for free or fee.
+## Target Audience
+
+The course is intended for researchers who want to learn coding for the first time with a data science application, or have explored programming and want to focus on fundamentals.
-- The material for this course can be viewed without login requirement on this [Bookdown website](LINK HERE). This format might be most appropriate for you if you rely on screen-reader technology.
-- This course can be taken for [free certification through Leanpub](LINK HERE).
-- This course can be taken on [Coursera for certification here](LINK HERE) (but it is not available for free on Coursera).
-- Our courses are open source, you can find the [source material for this course on GitHub](LINK HERE).
diff --git a/docs/no_toc/intro-to-computing.html b/docs/no_toc/intro-to-computing.html
index 6027c57..f08fd33 100644
--- a/docs/no_toc/intro-to-computing.html
+++ b/docs/no_toc/intro-to-computing.html
@@ -4,11 +4,11 @@
- Chapter 2 Intro to Computing | Introduction-to-R,-Season-1
-
+ Chapter 2 Intro to Computing | Season 1 Introduction to R
+
-
+
@@ -16,7 +16,7 @@
-
+
@@ -28,8 +28,8 @@
-
-
+
+
@@ -134,10 +134,10 @@
-
Slides that go with this lesson can be found here.
2.1 Goals of the course
@@ -305,35 +310,39 @@
2.9 Grammar Structure 2: Storing
To build up a computer program, we need to store our returned data type from our expression somewhere for downstream use. We can assign a variable to it as follows:
x =18+21
If you enter this in the Console, you will see that in the Environment, the variable x has a value of 39.
-
-
-
2.10 Execution rule for variable assignment
+
+
2.9.1 Execution rule for variable assignment
+
Evaluate the expression to the right of =.
Bind variable to the left of = to the resulting value.
The variable is stored in the environment.
<- is okay too!
-
+
The environment is where all the variables are stored, and can be used for an expression anytime once it is defined. Only one unique variable name can be defined.
The variable is stored in the working memory of your computer, Random Access Memory (RAM). This is temporary memory storage on the computer that can be accessed quickly. Typically a personal computer has 8, 16, 32 Gigabytes of RAM. When we work with large datasets, if you assign a variable to a data type larger than the available RAM, it will not work. More on this later.
Look, now x can be reused downstream:
x -2
## [1] 37
y = x *2
-
-
2.11 Grammar Structure 3: Evaluation of Functions
-
A function has a function name, arguments, and returns a data type.
-
-
2.12 Execution rule for functions:
+
+
+
2.10 Grammar Structure 3: Evaluation of Functions
+
A function has a function name, arguments, and returns a data type.
+
+
2.10.1 Execution rule for functions:
+
Evaluate the function by its arguments, and if the arguments are functions or contains operations, evaluate those functions or operations first.
The output of functions is called the returned value.
-
+
sqrt(nchar("hello"))
## [1] 2.236068
(nchar("hello") +4) *2
## [1] 18
-
-
2.13 Functions to read in data
+
+
+
+
2.11 Functions to read in data
We are going to read in a Comma Separated Value (CSV) spreadsheet, that contains information about cancer cell lines.
The first line calls the function read.csv() with a string argument representing the file path to the CSV file (we are using an URL online, but this is typically done locally), and the returned data type is stored in metadata variable. The resulting metadata variable is a new data type you have never seen before. It is a data structure called a data frame that we will be exploring next week. It holds a table of several data types that we can explore.
We run a few functions on metadata.
@@ -402,8 +411,8 @@
2.13 Functions to read in dataIf you don’t know what a function does, ask for help:
diff --git a/docs/no_toc/search_index.json b/docs/no_toc/search_index.json
index ccbf508..e5ba032 100644
--- a/docs/no_toc/search_index.json
+++ b/docs/no_toc/search_index.json
@@ -1 +1 @@
-[["introduction.html", "Chapter 1 Introduction 1.1 Target Audience 1.2 Curriculum", " Chapter 1 Introduction 1.1 Target Audience The course is intended for researchers who want to learn coding for the first time with a data science application, or have explored programming and want to focus on fundamentals. 1.2 Curriculum The course covers fundamentals of R, a high-level programming language, and use it to wrangle data for analysis and visualization. The programming skills you will learn are transferable to learn more about R independently and other high-level languages such as Python. At the end of the class, you will be reproducing analysis from a scientific publication! "],["intro-to-computing.html", "Chapter 2 Intro to Computing 2.1 Goals of the course 2.2 What is a computer program? 2.3 A programming language has following elements: 2.4 What is R and why should I use it? 2.5 R vs. Python as a first language 2.6 Posit Cloud Setup 2.7 Using Quarto for your work 2.8 Grammar Structure 1: Evaluation of Expressions 2.9 Grammar Structure 2: Storing data types in the global environment 2.10 Execution rule for variable assignment 2.11 Grammar Structure 3: Evaluation of Functions 2.12 Execution rule for functions: 2.13 Functions to read in data 2.14 Tips on Exercises / Debugging", " Chapter 2 Intro to Computing 2.1 Goals of the course Fundamental concepts in high-level programming languages (R, Python, Julia, WDL, etc.) that is transferable: How do programs run, and how do we solve problems using functions and data structures? Beginning of data science fundamentals: How do you translate your scientific question to a data wrangling problem and answer it? Data science workflow Find a nice balance between the two throughout the course: we will try to reproduce a figure from a scientific publication using new data. 2.2 What is a computer program? A sequence of instructions to manipulate data for the computer to execute. A series of translations: English <-> Programming Code for Interpreter <-> Machine Code for Central Processing Unit (CPU) We will focus on English <-> Programming Code for R Interpreter in this class. More importantly: How we organize ideas <-> Instructing a computer to do something. 2.3 A programming language has following elements: Grammar structure (simple building blocks) Means of combination to analyze and create content (examples around genomics provided, and your scientific creativity is strongly encouraged!) Means of abstraction for modular and reusable content (data structures, functions) Culture (emphasis on open-source, collaborative, reproducible code) Requires a lot of practice to be fluent! 2.4 What is R and why should I use it? It is a: Dynamic programming interpreter Highly used for data science, visualization, statistics, bioinformatics Open-source and free; easy to create and distribute your content; quirky culture 2.5 R vs. Python as a first language In terms of our goals, recall: Fundamental concepts in high-level programming languages Beginning of data science fundamentals There are a lot of nuances and debates, but I argue that Python is a better learning environment for the former and R is better for the latter. Ultimately, either should be okay! Perhaps more importantly, consider what your research group and collaborator are more comfortable with. 2.6 Posit Cloud Setup Posit Cloud/RStudio is an Integrated Development Environment (IDE). Think about it as Microsoft Word to a plain text editor. It provides extra bells and whistles to using R that is easier for the user. Today, we will pay close attention to: Script editor: where sequence of instructions are typed and saved as a text document as a R program. To run the program, the console will execute every single line of code in the document. Console (interpreter): Instead of giving a entire program in a text file, you could interact with the R Console line by line. You give it one line of instruction, and the console executes that single line. It is what R looks like without RStudio. Environment: Often, code will store information in memory, and it is shown in the environment. More on this later. 2.7 Using Quarto for your work Why should we use Quarto for data science work? Encourages reproducible workflows Code, output from code, and prose combined together Extendability to Python, Julia, and more. More options and guides can be found in Introduction to Quarto . 2.8 Grammar Structure 1: Evaluation of Expressions Expressions are be built out of operations or functions. Operations and functions combine data types to return another data type. We can combine multiple expressions together to form more complex expressions: an expression can have other expressions nested inside it. For instance, consider the following expressions entered to the R Console: 18 + 21 ## [1] 39 max(18, 21) ## [1] 21 max(18 + 21, 65) ## [1] 65 18 + (21 + 65) ## [1] 104 length("ATCG") ## [1] 1 Here, our input data types to the operation are numeric in lines 1-4 and our input data type to the function is character in line 5. Operations are just functions in hiding. We could have written: sum(18, 21) ## [1] 39 sum(18, sum(21, 65)) ## [1] 104 Remember the function machine from algebra class? We will use this schema to think about expressions. Function machine from algebra class. If an expression is made out of multiple, nested operations, what is the proper way of the R Console interpreting it? Being able to read nested operations and nested functions as a programmer is very important. 3 * 4 + 2 ## [1] 14 3 * (4 + 2) ## [1] 18 Lastly, a note on the use of functions: a programmer should not need to know how the function is implemented in order to use it - this emphasizes abstraction and modular thinking, a foundation in any programming language. 2.9 Grammar Structure 2: Storing data types in the global environment To build up a computer program, we need to store our returned data type from our expression somewhere for downstream use. We can assign a variable to it as follows: x = 18 + 21 If you enter this in the Console, you will see that in the Environment, the variable x has a value of 39. 2.10 Execution rule for variable assignment Evaluate the expression to the right of =. Bind variable to the left of = to the resulting value. The variable is stored in the environment. <- is okay too! The environment is where all the variables are stored, and can be used for an expression anytime once it is defined. Only one unique variable name can be defined. The variable is stored in the working memory of your computer, Random Access Memory (RAM). This is temporary memory storage on the computer that can be accessed quickly. Typically a personal computer has 8, 16, 32 Gigabytes of RAM. When we work with large datasets, if you assign a variable to a data type larger than the available RAM, it will not work. More on this later. Look, now x can be reused downstream: x - 2 ## [1] 37 y = x * 2 2.11 Grammar Structure 3: Evaluation of Functions A function has a function name, arguments, and returns a data type. 2.12 Execution rule for functions: Evaluate the function by its arguments, and if the arguments are functions or contains operations, evaluate those functions or operations first. The output of functions is called the returned value. sqrt(nchar("hello")) ## [1] 2.236068 (nchar("hello") + 4) * 2 ## [1] 18 2.13 Functions to read in data We are going to read in a Comma Separated Value (CSV) spreadsheet, that contains information about cancer cell lines. The first line calls the function read.csv() with a string argument representing the file path to the CSV file (we are using an URL online, but this is typically done locally), and the returned data type is stored in metadata variable. The resulting metadata variable is a new data type you have never seen before. It is a data structure called a data frame that we will be exploring next week. It holds a table of several data types that we can explore. We run a few functions on metadata. metadata = read.csv("https://github.com/caalo/Intro_to_R/raw/main/classroom_data/CCLE_metadata.csv") head(metadata) ## ModelID PatientID CellLineName StrippedCellLineName Age SourceType ## 1 ACH-000001 PT-gj46wT NIH:OVCAR-3 NIHOVCAR3 60 Commercial ## 2 ACH-000002 PT-5qa3uk HL-60 HL60 36 Commercial ## 3 ACH-000003 PT-puKIyc CACO2 CACO2 72 Commercial ## 4 ACH-000004 PT-q4K2cp HEL HEL 30 Commercial ## 5 ACH-000005 PT-q4K2cp HEL 92.1.7 HEL9217 30 Commercial ## 6 ACH-000006 PT-ej13Dz MONO-MAC-6 MONOMAC6 64 Commercial ## SangerModelID RRID DepmapModelType AgeCategory GrowthPattern ## 1 SIDM00105 CVCL_0465 HGSOC Adult Adherent ## 2 SIDM00829 CVCL_0002 AML Adult Suspension ## 3 SIDM00891 CVCL_0025 COAD Adult Adherent ## 4 SIDM00594 CVCL_0001 AML Adult Suspension ## 5 SIDM00593 CVCL_2481 AML Adult Mixed ## 6 SIDM01023 CVCL_1426 AMOL Adult Suspension ## LegacyMolecularSubtype PrimaryOrMetastasis SampleCollectionSite ## 1 Metastatic ascites ## 2 Primary haematopoietic_and_lymphoid_tissue ## 3 Primary Colon ## 4 Primary haematopoietic_and_lymphoid_tissue ## 5 bone_marrow ## 6 Primary haematopoietic_and_lymphoid_tissue ## Sex SourceDetail LegacySubSubtype CatalogNumber ## 1 Female ATCC high_grade_serous HTB-71 ## 2 Female ATCC M3 CCL-240 ## 3 Male ATCC HTB-37 ## 4 Male DSMZ M6 ACC 11 ## 5 Male ATCC M6 HEL9217 ## 6 Male DSMZ M5 ACC 124 ## CCLEName COSMICID PublicComments ## 1 NIHOVCAR3_OVARY 905933 ## 2 HL60_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 905938 ## 3 CACO2_LARGE_INTESTINE NA ## 4 HEL_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 907053 ## 5 HEL9217_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE NA ## 6 MONOMAC6_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 908148 ## WTSIMasterCellID EngineeredModel TreatmentStatus OnboardedMedia PlateCoating ## 1 2201 MF-001-041 None ## 2 55 MF-005-001 None ## 3 NA Unknown MF-015-009 None ## 4 783 Post-treatment MF-001-001 None ## 5 NA MF-001-001 None ## 6 2167 MF-001-001 None ## OncotreeCode OncotreeSubtype OncotreePrimaryDisease ## 1 HGSOC High-Grade Serous Ovarian Cancer Ovarian Epithelial Tumor ## 2 AML Acute Myeloid Leukemia Acute Myeloid Leukemia ## 3 COAD Colon Adenocarcinoma Colorectal Adenocarcinoma ## 4 AML Acute Myeloid Leukemia Acute Myeloid Leukemia ## 5 AML Acute Myeloid Leukemia Acute Myeloid Leukemia ## 6 AMOL Acute Monoblastic/Monocytic Leukemia Acute Myeloid Leukemia ## OncotreeLineage ## 1 Ovary/Fallopian Tube ## 2 Myeloid ## 3 Bowel ## 4 Myeloid ## 5 Myeloid ## 6 Myeloid nrow(metadata) ## [1] 1864 ncol(metadata) ## [1] 30 If you don’t know what a function does, ask for help: ?nrow 2.14 Tips on Exercises / Debugging Common errors: Syntax error. Changing a variable without realizing you did so. The function or operation does not accept the input data type. It did something else than I expected! Solutions: Where is the problem? What kind of problem is it? Explain your problem to someone! "],["about-the-authors.html", "About the Authors", " About the Authors These credits are based on our course contributors table guidelines. Credits Names Pedagogy Lead Content Instructor(s) FirstName LastName Lecturer(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved Delivered the course in some way - video or audio Content Author(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved If any other authors besides lead instructor Content Contributor(s) (include section name/link in parentheses) - make new line if more than one section involved Wrote less than a chapter Content Editor(s)/Reviewer(s) Checked your content Content Director(s) Helped guide the content direction Content Consultants (include chapter name/link in parentheses or word “General”) - make new line if more than one chapter involved Gave high level advice on content Acknowledgments Gave small assistance to content but not to the level of consulting Production Content Publisher(s) Helped with publishing platform Content Publishing Reviewer(s) Reviewed overall content and aesthetics on publishing platform Technical Course Publishing Engineer(s) Helped with the code for the technical aspects related to the specific course generation Template Publishing Engineers Candace Savonen, Carrie Wright, Ava Hoffman Publishing Maintenance Engineer Candace Savonen Technical Publishing Stylists Carrie Wright, Ava Hoffman, Candace Savonen Package Developers (ottrpal) Candace Savonen, John Muschelli, Carrie Wright Art and Design Illustrator(s) Created graphics for the course Figure Artist(s) Created figures/plots for course Videographer(s) Filmed videos Videography Editor(s) Edited film Audiographer(s) Recorded audio Audiography Editor(s) Edited audio recordings Funding Funder(s) Institution/individual who funded course including grant number Funding Staff Staff members who help with funding ## ─ Session info ─────────────────────────────────────────────────────────────── ## setting value ## version R version 4.0.2 (2020-06-22) ## os Ubuntu 20.04.5 LTS ## system x86_64, linux-gnu ## ui X11 ## language (EN) ## collate en_US.UTF-8 ## ctype en_US.UTF-8 ## tz Etc/UTC ## date 2023-09-28 ## ## ─ Packages ─────────────────────────────────────────────────────────────────── ## package * version date lib source ## assertthat 0.2.1 2019-03-21 [1] RSPM (R 4.0.5) ## bookdown 0.24 2023-03-28 [1] Github (rstudio/bookdown@88bc4ea) ## bslib 0.4.2 2022-12-16 [1] CRAN (R 4.0.2) ## cachem 1.0.7 2023-02-24 [1] CRAN (R 4.0.2) ## callr 3.5.0 2020-10-08 [1] RSPM (R 4.0.2) ## cli 3.6.1 2023-03-23 [1] CRAN (R 4.0.2) ## crayon 1.3.4 2017-09-16 [1] RSPM (R 4.0.0) ## desc 1.2.0 2018-05-01 [1] RSPM (R 4.0.3) ## devtools 2.3.2 2020-09-18 [1] RSPM (R 4.0.3) ## digest 0.6.25 2020-02-23 [1] RSPM (R 4.0.0) ## ellipsis 0.3.1 2020-05-15 [1] RSPM (R 4.0.3) ## evaluate 0.20 2023-01-17 [1] CRAN (R 4.0.2) ## fansi 0.4.1 2020-01-08 [1] RSPM (R 4.0.0) ## fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.0.2) ## fs 1.5.0 2020-07-31 [1] RSPM (R 4.0.3) ## glue 1.4.2 2020-08-27 [1] RSPM (R 4.0.5) ## hms 0.5.3 2020-01-08 [1] RSPM (R 4.0.0) ## htmltools 0.5.5 2023-03-23 [1] CRAN (R 4.0.2) ## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.0.2) ## jsonlite 1.7.1 2020-09-07 [1] RSPM (R 4.0.2) ## knitr 1.33 2023-03-28 [1] Github (yihui/knitr@a1052d1) ## lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.0.2) ## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.0.2) ## memoise 2.0.1 2021-11-26 [1] CRAN (R 4.0.2) ## ottrpal 1.0.1 2023-03-28 [1] Github (jhudsl/ottrpal@151e412) ## pillar 1.9.0 2023-03-22 [1] CRAN (R 4.0.2) ## pkgbuild 1.1.0 2020-07-13 [1] RSPM (R 4.0.2) ## pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.0.3) ## pkgload 1.1.0 2020-05-29 [1] RSPM (R 4.0.3) ## prettyunits 1.1.1 2020-01-24 [1] RSPM (R 4.0.3) ## processx 3.4.4 2020-09-03 [1] RSPM (R 4.0.2) ## ps 1.4.0 2020-10-07 [1] RSPM (R 4.0.2) ## R6 2.4.1 2019-11-12 [1] RSPM (R 4.0.0) ## readr 1.4.0 2020-10-05 [1] RSPM (R 4.0.2) ## remotes 2.2.0 2020-07-21 [1] RSPM (R 4.0.3) ## rlang 1.1.0 2023-03-14 [1] CRAN (R 4.0.2) ## rmarkdown 2.10 2023-03-28 [1] Github (rstudio/rmarkdown@02d3c25) ## rprojroot 2.0.3 2022-04-02 [1] CRAN (R 4.0.2) ## sass 0.4.5 2023-01-24 [1] CRAN (R 4.0.2) ## sessioninfo 1.1.1 2018-11-05 [1] RSPM (R 4.0.3) ## stringi 1.5.3 2020-09-09 [1] RSPM (R 4.0.3) ## stringr 1.4.0 2019-02-10 [1] RSPM (R 4.0.3) ## testthat 3.0.1 2023-03-28 [1] Github (R-lib/testthat@e99155a) ## tibble 3.2.1 2023-03-20 [1] CRAN (R 4.0.2) ## usethis 1.6.3 2020-09-17 [1] RSPM (R 4.0.2) ## utf8 1.1.4 2018-05-24 [1] RSPM (R 4.0.3) ## vctrs 0.6.1 2023-03-22 [1] CRAN (R 4.0.2) ## withr 2.3.0 2020-09-22 [1] RSPM (R 4.0.2) ## xfun 0.26 2023-03-28 [1] Github (yihui/xfun@74c2a66) ## yaml 2.2.1 2020-02-01 [1] RSPM (R 4.0.3) ## ## [1] /usr/local/lib/R/site-library ## [2] /usr/local/lib/R/library "],["references.html", "Chapter 3 References", " Chapter 3 References "],["404.html", "Page not found", " Page not found The page you requested cannot be found (perhaps it was moved or renamed). You may want to try searching to find the page's new location, or use the table of contents to find the page you are looking for. "]]
+[["index.html", "Season 1 Introduction to R Chapter 1 About this Course 1.1 Curriculum 1.2 Target Audience", " Season 1 Introduction to R September, 2023 Chapter 1 About this Course 1.1 Curriculum The course covers fundamentals of R, a high-level programming language, and use it to wrangle data for analysis and visualization. 1.2 Target Audience The course is intended for researchers who want to learn coding for the first time with a data science application, or have explored programming and want to focus on fundamentals. "],["intro-to-computing.html", "Chapter 2 Intro to Computing 2.1 Goals of the course 2.2 What is a computer program? 2.3 A programming language has following elements: 2.4 What is R and why should I use it? 2.5 R vs. Python as a first language 2.6 Posit Cloud Setup 2.7 Using Quarto for your work 2.8 Grammar Structure 1: Evaluation of Expressions 2.9 Grammar Structure 2: Storing data types in the global environment 2.10 Grammar Structure 3: Evaluation of Functions 2.11 Functions to read in data 2.12 Tips on Exercises / Debugging", " Chapter 2 Intro to Computing Slides that go with this lesson can be found here. 2.1 Goals of the course Fundamental concepts in high-level programming languages (R, Python, Julia, WDL, etc.) that is transferable: How do programs run, and how do we solve problems using functions and data structures? Beginning of data science fundamentals: How do you translate your scientific question to a data wrangling problem and answer it? Data science workflow Find a nice balance between the two throughout the course: we will try to reproduce a figure from a scientific publication using new data. 2.2 What is a computer program? A sequence of instructions to manipulate data for the computer to execute. A series of translations: English <-> Programming Code for Interpreter <-> Machine Code for Central Processing Unit (CPU) We will focus on English <-> Programming Code for R Interpreter in this class. More importantly: How we organize ideas <-> Instructing a computer to do something. 2.3 A programming language has following elements: Grammar structure (simple building blocks) Means of combination to analyze and create content (examples around genomics provided, and your scientific creativity is strongly encouraged!) Means of abstraction for modular and reusable content (data structures, functions) Culture (emphasis on open-source, collaborative, reproducible code) Requires a lot of practice to be fluent! 2.4 What is R and why should I use it? It is a: Dynamic programming interpreter Highly used for data science, visualization, statistics, bioinformatics Open-source and free; easy to create and distribute your content; quirky culture 2.5 R vs. Python as a first language In terms of our goals, recall: Fundamental concepts in high-level programming languages Beginning of data science fundamentals There are a lot of nuances and debates, but I argue that Python is a better learning environment for the former and R is better for the latter. Ultimately, either should be okay! Perhaps more importantly, consider what your research group and collaborator are more comfortable with. 2.6 Posit Cloud Setup Posit Cloud/RStudio is an Integrated Development Environment (IDE). Think about it as Microsoft Word to a plain text editor. It provides extra bells and whistles to using R that is easier for the user. Today, we will pay close attention to: Script editor: where sequence of instructions are typed and saved as a text document as a R program. To run the program, the console will execute every single line of code in the document. Console (interpreter): Instead of giving a entire program in a text file, you could interact with the R Console line by line. You give it one line of instruction, and the console executes that single line. It is what R looks like without RStudio. Environment: Often, code will store information in memory, and it is shown in the environment. More on this later. 2.7 Using Quarto for your work Why should we use Quarto for data science work? Encourages reproducible workflows Code, output from code, and prose combined together Extendability to Python, Julia, and more. More options and guides can be found in Introduction to Quarto . 2.8 Grammar Structure 1: Evaluation of Expressions Expressions are be built out of operations or functions. Operations and functions combine data types to return another data type. We can combine multiple expressions together to form more complex expressions: an expression can have other expressions nested inside it. For instance, consider the following expressions entered to the R Console: 18 + 21 ## [1] 39 max(18, 21) ## [1] 21 max(18 + 21, 65) ## [1] 65 18 + (21 + 65) ## [1] 104 length("ATCG") ## [1] 1 Here, our input data types to the operation are numeric in lines 1-4 and our input data type to the function is character in line 5. Operations are just functions in hiding. We could have written: sum(18, 21) ## [1] 39 sum(18, sum(21, 65)) ## [1] 104 Remember the function machine from algebra class? We will use this schema to think about expressions. Function machine from algebra class. If an expression is made out of multiple, nested operations, what is the proper way of the R Console interpreting it? Being able to read nested operations and nested functions as a programmer is very important. 3 * 4 + 2 ## [1] 14 3 * (4 + 2) ## [1] 18 Lastly, a note on the use of functions: a programmer should not need to know how the function is implemented in order to use it - this emphasizes abstraction and modular thinking, a foundation in any programming language. 2.9 Grammar Structure 2: Storing data types in the global environment To build up a computer program, we need to store our returned data type from our expression somewhere for downstream use. We can assign a variable to it as follows: x = 18 + 21 If you enter this in the Console, you will see that in the Environment, the variable x has a value of 39. 2.9.1 Execution rule for variable assignment Evaluate the expression to the right of =. Bind variable to the left of = to the resulting value. The variable is stored in the environment. <- is okay too! The environment is where all the variables are stored, and can be used for an expression anytime once it is defined. Only one unique variable name can be defined. The variable is stored in the working memory of your computer, Random Access Memory (RAM). This is temporary memory storage on the computer that can be accessed quickly. Typically a personal computer has 8, 16, 32 Gigabytes of RAM. When we work with large datasets, if you assign a variable to a data type larger than the available RAM, it will not work. More on this later. Look, now x can be reused downstream: x - 2 ## [1] 37 y = x * 2 2.10 Grammar Structure 3: Evaluation of Functions A function has a function name, arguments, and returns a data type. 2.10.1 Execution rule for functions: Evaluate the function by its arguments, and if the arguments are functions or contains operations, evaluate those functions or operations first. The output of functions is called the returned value. sqrt(nchar("hello")) ## [1] 2.236068 (nchar("hello") + 4) * 2 ## [1] 18 2.11 Functions to read in data We are going to read in a Comma Separated Value (CSV) spreadsheet, that contains information about cancer cell lines. The first line calls the function read.csv() with a string argument representing the file path to the CSV file (we are using an URL online, but this is typically done locally), and the returned data type is stored in metadata variable. The resulting metadata variable is a new data type you have never seen before. It is a data structure called a data frame that we will be exploring next week. It holds a table of several data types that we can explore. We run a few functions on metadata. metadata = read.csv("https://github.com/caalo/Intro_to_R/raw/main/classroom_data/CCLE_metadata.csv") head(metadata) ## ModelID PatientID CellLineName StrippedCellLineName Age SourceType ## 1 ACH-000001 PT-gj46wT NIH:OVCAR-3 NIHOVCAR3 60 Commercial ## 2 ACH-000002 PT-5qa3uk HL-60 HL60 36 Commercial ## 3 ACH-000003 PT-puKIyc CACO2 CACO2 72 Commercial ## 4 ACH-000004 PT-q4K2cp HEL HEL 30 Commercial ## 5 ACH-000005 PT-q4K2cp HEL 92.1.7 HEL9217 30 Commercial ## 6 ACH-000006 PT-ej13Dz MONO-MAC-6 MONOMAC6 64 Commercial ## SangerModelID RRID DepmapModelType AgeCategory GrowthPattern ## 1 SIDM00105 CVCL_0465 HGSOC Adult Adherent ## 2 SIDM00829 CVCL_0002 AML Adult Suspension ## 3 SIDM00891 CVCL_0025 COAD Adult Adherent ## 4 SIDM00594 CVCL_0001 AML Adult Suspension ## 5 SIDM00593 CVCL_2481 AML Adult Mixed ## 6 SIDM01023 CVCL_1426 AMOL Adult Suspension ## LegacyMolecularSubtype PrimaryOrMetastasis SampleCollectionSite ## 1 Metastatic ascites ## 2 Primary haematopoietic_and_lymphoid_tissue ## 3 Primary Colon ## 4 Primary haematopoietic_and_lymphoid_tissue ## 5 bone_marrow ## 6 Primary haematopoietic_and_lymphoid_tissue ## Sex SourceDetail LegacySubSubtype CatalogNumber ## 1 Female ATCC high_grade_serous HTB-71 ## 2 Female ATCC M3 CCL-240 ## 3 Male ATCC HTB-37 ## 4 Male DSMZ M6 ACC 11 ## 5 Male ATCC M6 HEL9217 ## 6 Male DSMZ M5 ACC 124 ## CCLEName COSMICID PublicComments ## 1 NIHOVCAR3_OVARY 905933 ## 2 HL60_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 905938 ## 3 CACO2_LARGE_INTESTINE NA ## 4 HEL_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 907053 ## 5 HEL9217_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE NA ## 6 MONOMAC6_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 908148 ## WTSIMasterCellID EngineeredModel TreatmentStatus OnboardedMedia PlateCoating ## 1 2201 MF-001-041 None ## 2 55 MF-005-001 None ## 3 NA Unknown MF-015-009 None ## 4 783 Post-treatment MF-001-001 None ## 5 NA MF-001-001 None ## 6 2167 MF-001-001 None ## OncotreeCode OncotreeSubtype OncotreePrimaryDisease ## 1 HGSOC High-Grade Serous Ovarian Cancer Ovarian Epithelial Tumor ## 2 AML Acute Myeloid Leukemia Acute Myeloid Leukemia ## 3 COAD Colon Adenocarcinoma Colorectal Adenocarcinoma ## 4 AML Acute Myeloid Leukemia Acute Myeloid Leukemia ## 5 AML Acute Myeloid Leukemia Acute Myeloid Leukemia ## 6 AMOL Acute Monoblastic/Monocytic Leukemia Acute Myeloid Leukemia ## OncotreeLineage ## 1 Ovary/Fallopian Tube ## 2 Myeloid ## 3 Bowel ## 4 Myeloid ## 5 Myeloid ## 6 Myeloid nrow(metadata) ## [1] 1864 ncol(metadata) ## [1] 30 If you don’t know what a function does, ask for help: ?nrow 2.12 Tips on Exercises / Debugging Common errors: Syntax error. Changing a variable without realizing you did so. The function or operation does not accept the input data type. It did something else than I expected! Solutions: Where is the problem? What kind of problem is it? Explain your problem to someone! "],["about-the-authors.html", "About the Authors", " About the Authors These credits are based on our course contributors table guidelines. Credits Names Pedagogy Lead Content Instructor(s) Chris Lo Lecturer Chris Lo Content Author(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved If any other authors besides lead instructor Content Contributor(s) (include section name/link in parentheses) - make new line if more than one section involved Wrote less than a chapter Content Editor(s)/Reviewer(s) Checked your content Content Director(s) Helped guide the content direction Content Consultants (include chapter name/link in parentheses or word “General”) - make new line if more than one chapter involved Gave high level advice on content Acknowledgments Gave small assistance to content but not to the level of consulting Production Content Publisher(s) Helped with publishing platform Content Publishing Reviewer(s) Reviewed overall content and aesthetics on publishing platform Technical Course Publishing Engineer(s) Helped with the code for the technical aspects related to the specific course generation Template Publishing Engineers Candace Savonen, Carrie Wright, Ava Hoffman Publishing Maintenance Engineer Candace Savonen Technical Publishing Stylists Carrie Wright, Ava Hoffman, Candace Savonen Package Developers (ottrpal) Candace Savonen, John Muschelli, Carrie Wright Art and Design Illustrator(s) Created graphics for the course Figure Artist(s) Created figures/plots for course Videographer(s) Filmed videos Videography Editor(s) Edited film Audiographer(s) Recorded audio Audiography Editor(s) Edited audio recordings Funding Funder(s) Institution/individual who funded course including grant number Funding Staff Staff members who help with funding ## ─ Session info ─────────────────────────────────────────────────────────────── ## setting value ## version R version 4.0.2 (2020-06-22) ## os Ubuntu 20.04.5 LTS ## system x86_64, linux-gnu ## ui X11 ## language (EN) ## collate en_US.UTF-8 ## ctype en_US.UTF-8 ## tz Etc/UTC ## date 2023-09-28 ## ## ─ Packages ─────────────────────────────────────────────────────────────────── ## package * version date lib source ## assertthat 0.2.1 2019-03-21 [1] RSPM (R 4.0.5) ## bookdown 0.24 2023-03-28 [1] Github (rstudio/bookdown@88bc4ea) ## bslib 0.4.2 2022-12-16 [1] CRAN (R 4.0.2) ## cachem 1.0.7 2023-02-24 [1] CRAN (R 4.0.2) ## callr 3.5.0 2020-10-08 [1] RSPM (R 4.0.2) ## cli 3.6.1 2023-03-23 [1] CRAN (R 4.0.2) ## crayon 1.3.4 2017-09-16 [1] RSPM (R 4.0.0) ## desc 1.2.0 2018-05-01 [1] RSPM (R 4.0.3) ## devtools 2.3.2 2020-09-18 [1] RSPM (R 4.0.3) ## digest 0.6.25 2020-02-23 [1] RSPM (R 4.0.0) ## ellipsis 0.3.1 2020-05-15 [1] RSPM (R 4.0.3) ## evaluate 0.20 2023-01-17 [1] CRAN (R 4.0.2) ## fansi 0.4.1 2020-01-08 [1] RSPM (R 4.0.0) ## fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.0.2) ## fs 1.5.0 2020-07-31 [1] RSPM (R 4.0.3) ## glue 1.4.2 2020-08-27 [1] RSPM (R 4.0.5) ## hms 0.5.3 2020-01-08 [1] RSPM (R 4.0.0) ## htmltools 0.5.5 2023-03-23 [1] CRAN (R 4.0.2) ## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.0.2) ## jsonlite 1.7.1 2020-09-07 [1] RSPM (R 4.0.2) ## knitr 1.33 2023-03-28 [1] Github (yihui/knitr@a1052d1) ## lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.0.2) ## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.0.2) ## memoise 2.0.1 2021-11-26 [1] CRAN (R 4.0.2) ## ottrpal 1.0.1 2023-03-28 [1] Github (jhudsl/ottrpal@151e412) ## pillar 1.9.0 2023-03-22 [1] CRAN (R 4.0.2) ## pkgbuild 1.1.0 2020-07-13 [1] RSPM (R 4.0.2) ## pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.0.3) ## pkgload 1.1.0 2020-05-29 [1] RSPM (R 4.0.3) ## prettyunits 1.1.1 2020-01-24 [1] RSPM (R 4.0.3) ## processx 3.4.4 2020-09-03 [1] RSPM (R 4.0.2) ## ps 1.4.0 2020-10-07 [1] RSPM (R 4.0.2) ## R6 2.4.1 2019-11-12 [1] RSPM (R 4.0.0) ## readr 1.4.0 2020-10-05 [1] RSPM (R 4.0.2) ## remotes 2.2.0 2020-07-21 [1] RSPM (R 4.0.3) ## rlang 1.1.0 2023-03-14 [1] CRAN (R 4.0.2) ## rmarkdown 2.10 2023-03-28 [1] Github (rstudio/rmarkdown@02d3c25) ## rprojroot 2.0.3 2022-04-02 [1] CRAN (R 4.0.2) ## sass 0.4.5 2023-01-24 [1] CRAN (R 4.0.2) ## sessioninfo 1.1.1 2018-11-05 [1] RSPM (R 4.0.3) ## stringi 1.5.3 2020-09-09 [1] RSPM (R 4.0.3) ## stringr 1.4.0 2019-02-10 [1] RSPM (R 4.0.3) ## testthat 3.0.1 2023-03-28 [1] Github (R-lib/testthat@e99155a) ## tibble 3.2.1 2023-03-20 [1] CRAN (R 4.0.2) ## usethis 1.6.3 2020-09-17 [1] RSPM (R 4.0.2) ## utf8 1.1.4 2018-05-24 [1] RSPM (R 4.0.3) ## vctrs 0.6.1 2023-03-22 [1] CRAN (R 4.0.2) ## withr 2.3.0 2020-09-22 [1] RSPM (R 4.0.2) ## xfun 0.26 2023-03-28 [1] Github (yihui/xfun@74c2a66) ## yaml 2.2.1 2020-02-01 [1] RSPM (R 4.0.3) ## ## [1] /usr/local/lib/R/site-library ## [2] /usr/local/lib/R/library "],["references.html", "Chapter 3 References", " Chapter 3 References "],["404.html", "Page not found", " Page not found The page you requested cannot be found (perhaps it was moved or renamed). You may want to try searching to find the page's new location, or use the table of contents to find the page you are looking for. "]]
diff --git a/docs/no_toc/slides/lesson1_slides.html b/docs/no_toc/slides/lesson1_slides.html
new file mode 100644
index 0000000..247e9a3
--- /dev/null
+++ b/docs/no_toc/slides/lesson1_slides.html
@@ -0,0 +1,1242 @@
+
+
+
+
+
+
+
+
+
+
+
+
+ W1: Intro to Computing
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
W1: Intro to Computing
+
+
+
+
+
+
+
Introductions
+
+
Who am I?
+
+
+
+
What is DaSL?
+
+
+
+
+
Who are you?
+
+
Name, pronouns, group you work in
+
What you want to get out of the class
+
Favorite fall activity
+
+
+
+
+
+
Goals of the course
+
+
+
Fundamental concepts in high-level programming languages: How do programs run, and how do we solve problems using functions and data structures?
+
+
+
+
+
Beginning of data science fundamentals: How do you translate your scientific question to a data wrangling problem and answer it?
+
+
+
+Data science workflow
+
+
+
+
+
+
+
Culture of the course
+
+
+
Learning on the job is challenging
+
+
I will move at learner’s pace; collaborative learning encouraged
+
+
+
+
+
+
Various exposure to programming and data science
+
+
Take what you need: some will be new, some will be familiar
+
+
+
+
+
+
Various personal goals and applications
+
+
Curate content towards end of the course
+
+
+
+
+
+
Respect Code of Conduct
+
+
+
+
+
Format of the course
+
+
+
6 classes, come to 5 of them in-person if you can
+
+
+
+
+
Streamed online, recordings will be available
+
+
+
+
+
1-2 hour exercises after each session are strongly encouraged as they provide practice and preview the next class
+
+
+
+
+
Online discussion on Slack
+
+
+
+
+
Certification quiz at the end of the course
+
+
+
+
+
What is a computer program?
+
+
+
A sequence of instructions to manipulate data for the computer to execute.
+
+
+
+
+
A series of translations: English <-> Programming Code for Interpreter <-> Machine Code for Central Processing Unit (CPU)
+
+
+
+
We will focus on English <-> Programming Code for R Interpreter in this class.
+
+
+
More importantly: How we organize ideas <-> Instructing a computer to do something.
+
+
+
+
A programming language has following themes:
+
+
+
Grammar structure (simple building blocks)
+
+
+
+
+
Means of combination to analyze and create content (examples around genomics)
+
+
+
+
+
Means of abstraction for modular and reusable content (data structures, functions)
+
+
+
+
+
Culture (emphasis on open-source, collaborative, reproducible code)
+
+
+
+
+
What is R and why should I use it?
+
+
+
Dynamic programming interpreter
+
+
+
+
+
Highly used for data science, visualization, statistics, bioinformatics
+
+
+
+
+
Open-source and free; easy to create and distribute your content; kind, quirky culture
+
+
+
+
+
Setting up Posit Cloud and trying out your first analysis!
+
link here
+
+
+
Break
+
+
+
Grammar Structure 1: Evaluation of Expressions
+
+
+
Expressions are be built out of operations or functions.
+
+
+
+
+
Operations and functions combine data types to return another data type.
+
+
+
+
+
We can combine multiple expressions together to form more complex expressions: an expression can have other expressions nested inside it.
+
+
+
+
+
Examples
+
+
18+21
+
+
+
+
[1] 39
+
+
+
+
+
max(18, 21)
+
+
+
+
[1] 21
+
+
+
+
+
+
max(18+21, 65)
+
+
+
+
[1] 65
+
+
+
+
+
+
18+ (21+65)
+
+
+
+
[1] 104
+
+
+
+
+
+
length("ATCG")
+
+
+
+
[1] 1
+
+
+
+
+
+
+
Data types
+
+
Numeric: 18, 21, 65, 1.25
+
Character: “ATCG”, “Whatever”, “948-293-0000”
+
Logical: TRUE, FALSE
+
+
+
+
Function machine from algebra class
+
+
+
+
+
Operations are just functions. We could have written:
+
+
sum(18, 21)
+
+
+
+
[1] 39
+
+
+
+
+
+
sum(18, sum(21, 65))
+
+
+
+
[1] 104
+
+
+
+
+
+
+
Grammar Structure 2: Storing data types in the global environment
+
+
To build up a computer program, we need to store our returned data type from our expression somewhere for downstream use.
+
+
x =18+21
+
+
+
+
+
+
+
+
+
+
+
+
+
Execution rule for variable assignment
+
+
+
Evaluate the expression to the right of =.
+
Bind variable to the left of = to the resulting value.
+
The variable is stored in the environment.
+
<- is okay too!
+
+
+
+
+
+
+
+
Downstream
+
Look, now x can be reused downstream:
+
+
x -2
+
+
+
+
[1] 37
+
+
+
+
+
y = x *2
+y
+
+
+
+
[1] 78
+
+
+
+
+
+
Grammar Structure 3: Evaluation of Functions
+
A function has a function name, arguments, and returns a data type.
+
+
+
+
+
+
+
+
Execution rule for functions:
+
+
+
Evaluate the function by its arguments, and if the arguments are functions or contains operations, evaluate those functions or operations first.
+
The output of functions is called the returned value.
+
+
+
+
+
+
+
sqrt(nchar("hello"))
+
+
+
+
[1] 2.236068
+
+
+
+
+
+
(nchar("hello") +4) *2
+
+
+
+
[1] 18
+
+
+
+
+
+
Tips on Exercises / Debugging
+
+
Common errors:
+
+
Syntax error.
+
+
+
+
+
The function or operation does not accept the input data type.
+
+
+
+
+
Changing a variable without realizing you did so.
+
+
+
+
+
It did something else than I expected!
+
+
+
+
+
Solutions:
+
+
Where is the problem?
+
+
+
+
What kind of problem is it?
+
+
+
+
+
Explain your problem to someone!
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
\ No newline at end of file