diff --git a/.github/workflows/test.yml b/.github/workflows/test.yml
index d22022e..1e6aa40 100644
--- a/.github/workflows/test.yml
+++ b/.github/workflows/test.yml
@@ -15,7 +15,7 @@ jobs:
fail-fast: true
matrix:
os: ["ubuntu-latest", "macos-latest", "windows-latest"]
- python-version: ["3.8", "3.9", "3.10"]
+ python-version: ["3.8", "3.9", "3.10", "3.11"]
steps:
- name: Checkout
uses: actions/checkout@v3
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
index 88e81b0..892b0c7 100644
--- a/.pre-commit-config.yaml
+++ b/.pre-commit-config.yaml
@@ -55,7 +55,7 @@ repos:
- id: black
# - id: black-jupyter
- repo: https://github.com/charliermarsh/ruff-pre-commit
- rev: v0.0.263
+ rev: v0.0.264
hooks:
- id: ruff
args: [--fix-only, --show-fixes]
@@ -81,7 +81,7 @@ repos:
additional_dependencies: [tomli]
files: ^(graphblas_algorithms|docs)/
- repo: https://github.com/charliermarsh/ruff-pre-commit
- rev: v0.0.263
+ rev: v0.0.264
hooks:
- id: ruff
# `pyroma` may help keep our package standards up to date if best practices change.
diff --git a/MANIFEST.in b/MANIFEST.in
index 92306c0..c69947d 100644
--- a/MANIFEST.in
+++ b/MANIFEST.in
@@ -4,6 +4,3 @@ include setup.py
include README.md
include LICENSE
include MANIFEST.in
-docs/_static/img/logo-name-medium.png
-docs/_static/img/graphblas-vs-igraph.png
-docs/_static/img/graphblas-vs-networkx.png
diff --git a/README.md b/README.md
index 32039fb..821cd95 100644
--- a/README.md
+++ b/README.md
@@ -1,4 +1,4 @@
-
+
[](https://anaconda.org/conda-forge/graphblas-algorithms)
[](https://pypi.python.org/pypi/graphblas-algorithms/)
@@ -21,9 +21,9 @@ Why use GraphBLAS Algorithms? Because it is *fast*, *flexible*, and *familiar* b
Are we missing any [algorithms](#Plugin-Algorithms) that you want?
[Please let us know!](https://github.com/python-graphblas/graphblas-algorithms/issues)
- +
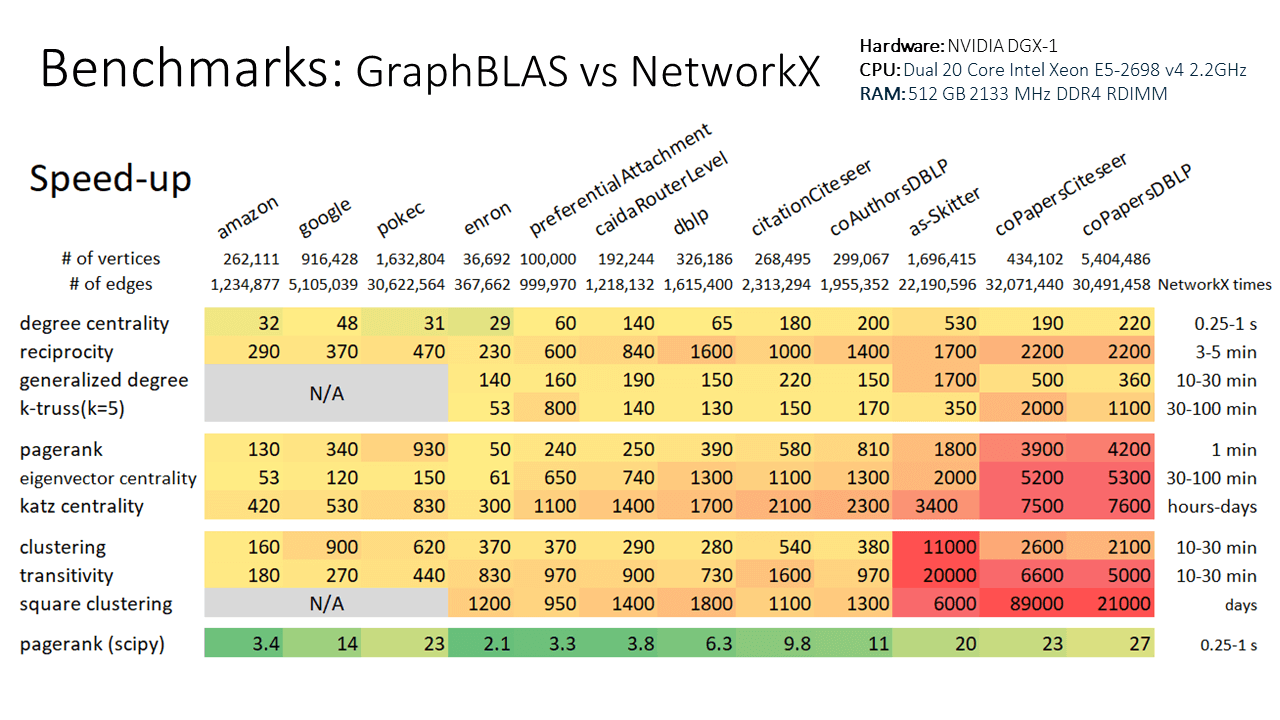
+
- +
+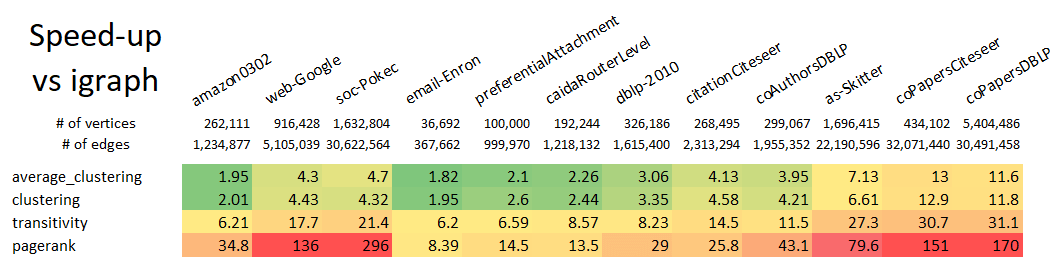 ### Installation
```
@@ -151,6 +151,8 @@ dispatch pattern shown above.
- descendants
- Dominating
- is_dominating_set
+- Generators
+ - ego_graph
- Isolate
- is_isolate
- isolates
diff --git a/graphblas_algorithms/__init__.py b/graphblas_algorithms/__init__.py
index f9bbcf4..09418f5 100644
--- a/graphblas_algorithms/__init__.py
+++ b/graphblas_algorithms/__init__.py
@@ -3,6 +3,7 @@
from .classes import *
from .algorithms import * # isort:skip
+from .generators import * # isort:skip
try:
__version__ = importlib.metadata.version("graphblas-algorithms")
diff --git a/graphblas_algorithms/algorithms/_bfs.py b/graphblas_algorithms/algorithms/_bfs.py
new file mode 100644
index 0000000..31674ce
--- /dev/null
+++ b/graphblas_algorithms/algorithms/_bfs.py
@@ -0,0 +1,150 @@
+"""BFS routines used by other algorithms"""
+
+import numpy as np
+from graphblas import Matrix, Vector, binary, replace, unary
+from graphblas.semiring import any_pair
+
+
+def _get_cutoff(n, cutoff):
+ if cutoff is None or cutoff >= n:
+ return n # Everything
+ return cutoff + 1 # Inclusive
+
+
+def _plain_bfs(G, source, *, cutoff=None):
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q = Vector(bool, n, name="q")
+ v[index] = True
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for _i in range(1, cutoff):
+ q(~v.S, replace) << any_pair_bool(q @ A)
+ if q.nvals == 0:
+ break
+ v(q.S) << True
+ return v
+
+
+def _bfs_level(G, source, cutoff=None, *, transpose=False, dtype=int):
+ if dtype == bool:
+ dtype = int
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ if transpose and G.is_directed():
+ A = A.T # TODO: should we use "AT" instead?
+ n = A.nrows
+ v = Vector(dtype, n, name="bfs_level")
+ q = Vector(bool, n, name="q")
+ v[index] = 0
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for i in range(1, cutoff):
+ q(~v.S, replace) << any_pair_bool(q @ A)
+ if q.nvals == 0:
+ break
+ v(q.S) << i
+ return v
+
+
+def _bfs_levels(G, nodes, cutoff=None, *, dtype=int):
+ if dtype == bool:
+ dtype = int
+ A = G.get_property("offdiag")
+ n = A.nrows
+ if nodes is None:
+ # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
+ D = Vector(dtype, n, name="bfs_levels_vector")
+ D << 0
+ D = D.diag(name="bfs_levels")
+ else:
+ ids = G.list_to_ids(nodes)
+ D = Matrix.from_coo(
+ np.arange(len(ids), dtype=np.uint64),
+ ids,
+ 0,
+ dtype,
+ nrows=len(ids),
+ ncols=n,
+ name="bfs_levels",
+ )
+ Q = unary.one[bool](D).new(name="Q")
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for i in range(1, cutoff):
+ Q(~D.S, replace) << any_pair_bool(Q @ A)
+ if Q.nvals == 0:
+ break
+ D(Q.S) << i
+ return D
+
+
+# TODO: benchmark this and the version commented out below
+def _plain_bfs_bidirectional(G, source):
+ # Bi-directional BFS w/o symmetrizing the adjacency matrix
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ # XXX: should we use `AT` if available?
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q_out = Vector(bool, n, name="q_out")
+ q_in = Vector(bool, n, name="q_in")
+ v[index] = True
+ q_in[index] = True
+ any_pair_bool = any_pair[bool]
+ is_out_empty = True
+ is_in_empty = False
+ for _i in range(1, n):
+ # Traverse out-edges from the most recent `q_in` and `q_out`
+ if is_out_empty:
+ q_out(~v.S) << any_pair_bool(q_in @ A)
+ else:
+ q_out << binary.any(q_out | q_in)
+ q_out(~v.S, replace) << any_pair_bool(q_out @ A)
+ is_out_empty = q_out.nvals == 0
+ if not is_out_empty:
+ v(q_out.S) << True
+ elif is_in_empty:
+ break
+ # Traverse in-edges from the most recent `q_in` and `q_out`
+ if is_in_empty:
+ q_in(~v.S) << any_pair_bool(A @ q_out)
+ else:
+ q_in << binary.any(q_out | q_in)
+ q_in(~v.S, replace) << any_pair_bool(A @ q_in)
+ is_in_empty = q_in.nvals == 0
+ if not is_in_empty:
+ v(q_in.S) << True
+ elif is_out_empty:
+ break
+ return v
+
+
+"""
+def _plain_bfs_bidirectional(G, source):
+ # Bi-directional BFS w/o symmetrizing the adjacency matrix
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q = Vector(bool, n, name="q")
+ q2 = Vector(bool, n, name="q_2")
+ v[index] = True
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ for _i in range(1, n):
+ q2(~v.S, replace) << any_pair_bool(q @ A)
+ v(q2.S) << True
+ q(~v.S, replace) << any_pair_bool(A @ q)
+ if q.nvals == 0:
+ if q2.nvals == 0:
+ break
+ q, q2 = q2, q
+ elif q2.nvals != 0:
+ q << binary.any(q | q2)
+ return v
+"""
diff --git a/graphblas_algorithms/algorithms/centrality/eigenvector.py b/graphblas_algorithms/algorithms/centrality/eigenvector.py
index 5172f61..e9385f3 100644
--- a/graphblas_algorithms/algorithms/centrality/eigenvector.py
+++ b/graphblas_algorithms/algorithms/centrality/eigenvector.py
@@ -1,11 +1,7 @@
from graphblas import Vector
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import (
- ConvergenceFailure,
- GraphBlasAlgorithmException,
- PointlessConcept,
-)
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure, GraphBlasAlgorithmException, PointlessConcept
__all__ = ["eigenvector_centrality"]
diff --git a/graphblas_algorithms/algorithms/centrality/katz.py b/graphblas_algorithms/algorithms/centrality/katz.py
index 78de982..8087e85 100644
--- a/graphblas_algorithms/algorithms/centrality/katz.py
+++ b/graphblas_algorithms/algorithms/centrality/katz.py
@@ -2,11 +2,8 @@
from graphblas.core.utils import output_type
from graphblas.semiring import plus_first, plus_times
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import (
- ConvergenceFailure,
- GraphBlasAlgorithmException,
-)
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure, GraphBlasAlgorithmException
__all__ = ["katz_centrality"]
diff --git a/graphblas_algorithms/algorithms/components/connected.py b/graphblas_algorithms/algorithms/components/connected.py
index 37c0fc9..fb2f678 100644
--- a/graphblas_algorithms/algorithms/components/connected.py
+++ b/graphblas_algorithms/algorithms/components/connected.py
@@ -1,7 +1,5 @@
-from graphblas import Vector, replace
-from graphblas.semiring import any_pair
-
-from graphblas_algorithms.algorithms.exceptions import PointlessConcept
+from .._bfs import _plain_bfs
+from ..exceptions import PointlessConcept
def is_connected(G):
@@ -12,20 +10,3 @@ def is_connected(G):
def node_connected_component(G, n):
return _plain_bfs(G, n)
-
-
-def _plain_bfs(G, source):
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q = Vector(bool, n, name="q")
- v[index] = True
- q[index] = True
- any_pair_bool = any_pair[bool]
- for _i in range(1, n):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << True
- return v
diff --git a/graphblas_algorithms/algorithms/components/weakly_connected.py b/graphblas_algorithms/algorithms/components/weakly_connected.py
index eb3dc75..99eba78 100644
--- a/graphblas_algorithms/algorithms/components/weakly_connected.py
+++ b/graphblas_algorithms/algorithms/components/weakly_connected.py
@@ -1,77 +1,8 @@
-from graphblas import Vector, binary, replace
-from graphblas.semiring import any_pair
-
-from graphblas_algorithms.algorithms.exceptions import PointlessConcept
+from .._bfs import _plain_bfs_bidirectional
+from ..exceptions import PointlessConcept
def is_weakly_connected(G):
if len(G) == 0:
raise PointlessConcept("Connectivity is undefined for the null graph.")
- return _plain_bfs(G, next(iter(G))).nvals == len(G)
-
-
-# TODO: benchmark this and the version commented out below
-def _plain_bfs(G, source):
- # Bi-directional BFS w/o symmetrizing the adjacency matrix
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- # XXX: should we use `AT` if available?
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q_out = Vector(bool, n, name="q_out")
- q_in = Vector(bool, n, name="q_in")
- v[index] = True
- q_in[index] = True
- any_pair_bool = any_pair[bool]
- is_out_empty = True
- is_in_empty = False
- for _i in range(1, n):
- # Traverse out-edges from the most recent `q_in` and `q_out`
- if is_out_empty:
- q_out(~v.S) << any_pair_bool(q_in @ A)
- else:
- q_out << binary.any(q_out | q_in)
- q_out(~v.S, replace) << any_pair_bool(q_out @ A)
- is_out_empty = q_out.nvals == 0
- if not is_out_empty:
- v(q_out.S) << True
- elif is_in_empty:
- break
- # Traverse in-edges from the most recent `q_in` and `q_out`
- if is_in_empty:
- q_in(~v.S) << any_pair_bool(A @ q_out)
- else:
- q_in << binary.any(q_out | q_in)
- q_in(~v.S, replace) << any_pair_bool(A @ q_in)
- is_in_empty = q_in.nvals == 0
- if not is_in_empty:
- v(q_in.S) << True
- elif is_out_empty:
- break
- return v
-
-
-"""
-def _plain_bfs(G, source):
- # Bi-directional BFS w/o symmetrizing the adjacency matrix
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q = Vector(bool, n, name="q")
- q2 = Vector(bool, n, name="q_2")
- v[index] = True
- q[index] = True
- any_pair_bool = any_pair[bool]
- for _i in range(1, n):
- q2(~v.S, replace) << any_pair_bool(q @ A)
- v(q2.S) << True
- q(~v.S, replace) << any_pair_bool(A @ q)
- if q.nvals == 0:
- if q2.nvals == 0:
- break
- q, q2 = q2, q
- elif q2.nvals != 0:
- q << binary.any(q | q2)
- return v
-"""
+ return _plain_bfs_bidirectional(G, next(iter(G))).nvals == len(G)
diff --git a/graphblas_algorithms/algorithms/core.py b/graphblas_algorithms/algorithms/core.py
index 8133c71..a6ff26d 100644
--- a/graphblas_algorithms/algorithms/core.py
+++ b/graphblas_algorithms/algorithms/core.py
@@ -1,11 +1,12 @@
from graphblas import Matrix, monoid, replace, select, semiring
-from graphblas_algorithms.classes.graph import Graph
+from graphblas_algorithms import Graph
__all__ = ["k_truss"]
def k_truss(G: Graph, k) -> Graph:
+ # TODO: should we have an option to keep the output matrix the same size?
# Ignore self-edges
S = G.get_property("offdiag")
@@ -32,6 +33,5 @@ def k_truss(G: Graph, k) -> Graph:
Ktruss = C[indices, indices].new()
# Convert back to networkx graph with correct node ids
- keys = G.list_to_keys(indices)
- key_to_id = dict(zip(keys, range(len(indices))))
+ key_to_id = G.renumber_key_to_id(indices.tolist())
return Graph(Ktruss, key_to_id=key_to_id)
diff --git a/graphblas_algorithms/algorithms/link_analysis/hits_alg.py b/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
index 515806e..662ac14 100644
--- a/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
+++ b/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
@@ -1,7 +1,7 @@
from graphblas import Vector
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import ConvergenceFailure
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure
__all__ = ["hits"]
diff --git a/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py b/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
index 518c09f..d665e98 100644
--- a/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
+++ b/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
@@ -3,8 +3,9 @@
from graphblas.semiring import plus_first, plus_times
from graphblas_algorithms import Graph
-from graphblas_algorithms.algorithms._helpers import is_converged
-from graphblas_algorithms.algorithms.exceptions import ConvergenceFailure
+
+from .._helpers import is_converged
+from ..exceptions import ConvergenceFailure
__all__ = ["pagerank", "google_matrix"]
diff --git a/graphblas_algorithms/algorithms/shortest_paths/unweighted.py b/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
index 3c8243f..5062e87 100644
--- a/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
+++ b/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
@@ -1,6 +1,6 @@
-import numpy as np
-from graphblas import Matrix, Vector, replace, unary
-from graphblas.semiring import any_pair
+from graphblas import Matrix
+
+from .._bfs import _bfs_level, _bfs_levels
__all__ = [
"single_source_shortest_path_length",
@@ -25,59 +25,3 @@ def all_pairs_shortest_path_length(G, cutoff=None, *, nodes=None, expand_output=
rv[ids, :] = D
return rv
return D
-
-
-def _bfs_level(G, source, cutoff, *, transpose=False):
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- if transpose and G.is_directed():
- A = A.T # TODO: should we use "AT" instead?
- n = A.nrows
- v = Vector(int, n, name="bfs_unweighted")
- q = Vector(bool, n, name="q")
- v[index] = 0
- q[index] = True
- any_pair_bool = any_pair[bool]
- if cutoff is None or cutoff >= n:
- cutoff = n # Everything
- else:
- cutoff += 1 # Inclusive
- for i in range(1, cutoff):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << i
- return v
-
-
-def _bfs_levels(G, nodes, cutoff):
- A = G.get_property("offdiag")
- n = A.nrows
- if nodes is None:
- # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
- D = Vector(int, n, name="bfs_unweighted_vector")
- D << 0
- D = D.diag(name="bfs_unweighted")
- else:
- ids = G.list_to_ids(nodes)
- D = Matrix.from_coo(
- np.arange(len(ids), dtype=np.uint64),
- ids,
- 0,
- int,
- nrows=len(ids),
- ncols=n,
- name="bfs_unweighted",
- )
- Q = unary.one[bool](D).new(name="Q")
- any_pair_bool = any_pair[bool]
- if cutoff is None or cutoff >= n:
- cutoff = n # Everything
- else:
- cutoff += 1 # Inclusive
- for i in range(1, cutoff):
- Q(~D.S, replace) << any_pair_bool(Q @ A)
- if Q.nvals == 0:
- break
- D(Q.S) << i
- return D
diff --git a/graphblas_algorithms/algorithms/shortest_paths/weighted.py b/graphblas_algorithms/algorithms/shortest_paths/weighted.py
index 8e6efef..fddf672 100644
--- a/graphblas_algorithms/algorithms/shortest_paths/weighted.py
+++ b/graphblas_algorithms/algorithms/shortest_paths/weighted.py
@@ -2,6 +2,7 @@
from graphblas import Matrix, Vector, binary, monoid, replace, select, unary
from graphblas.semiring import any_pair, min_plus
+from .._bfs import _bfs_level, _bfs_levels
from ..exceptions import Unbounded
__all__ = [
@@ -11,14 +12,16 @@
]
-def single_source_bellman_ford_path_length(G, source):
+def single_source_bellman_ford_path_length(G, source, *, cutoff=None):
# No need for `is_weighted=` keyword, b/c this is assumed to be weighted (I think)
index = G._key_to_id[source]
if G.get_property("is_iso"):
# If the edges are iso-valued (and positive), then we can simply do level BFS
is_negative, iso_value = G.get_properties("has_negative_edges+ iso_value")
if not is_negative:
- d = _bfs_level(G, source, dtype=iso_value.dtype)
+ if cutoff is not None:
+ cutoff = int(cutoff // iso_value)
+ d = _bfs_level(G, source, cutoff, dtype=iso_value.dtype)
if iso_value != 1:
d *= iso_value
return d
@@ -49,6 +52,8 @@ def single_source_bellman_ford_path_length(G, source):
# `cur` is the current frontier of values that improved in the previous iteration.
# This means that in this iteration we drop values from `cur` that are not better.
cur << min_plus(cur @ A)
+ if cutoff is not None:
+ cur << select.valuele(cur, cutoff)
# Mask is True where cur not in d or cur < d
mask << one(cur)
@@ -63,6 +68,8 @@ def single_source_bellman_ford_path_length(G, source):
else:
# Check for negative cycle when for loop completes without breaking
cur << min_plus(cur @ A)
+ if cutoff is not None:
+ cur << select.valuele(cur, cutoff)
mask << binary.lt(cur & d)
if mask.reduce(monoid.lor):
raise Unbounded("Negative cycle detected.")
@@ -157,56 +164,6 @@ def bellman_ford_path_lengths(G, nodes=None, *, expand_output=False):
return D
-def _bfs_level(G, source, *, dtype=int):
- if dtype == bool:
- dtype = int
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(dtype, n, name="bfs_level")
- q = Vector(bool, n, name="q")
- v[index] = 0
- q[index] = True
- any_pair_bool = any_pair[bool]
- for i in range(1, n):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << i
- return v
-
-
-def _bfs_levels(G, nodes=None, *, dtype=int):
- if dtype == bool:
- dtype = int
- A = G.get_property("offdiag")
- n = A.nrows
- if nodes is None:
- # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
- D = Vector(dtype, n, name="bfs_levels_vector")
- D << 0
- D = D.diag(name="bfs_levels")
- else:
- ids = G.list_to_ids(nodes)
- D = Matrix.from_coo(
- np.arange(len(ids), dtype=np.uint64),
- ids,
- 0,
- dtype,
- nrows=len(ids),
- ncols=n,
- name="bfs_levels",
- )
- Q = unary.one[bool](D).new(name="Q")
- any_pair_bool = any_pair[bool]
- for i in range(1, n):
- Q(~D.S, replace) << any_pair_bool(Q @ A)
- if Q.nvals == 0:
- break
- D(Q.S) << i
- return D
-
-
def negative_edge_cycle(G):
# TODO: use a heuristic to try to stop early
if G.is_directed():
diff --git a/graphblas_algorithms/algorithms/triads.py b/graphblas_algorithms/algorithms/triads.py
index 54702c7..e6ec2be 100644
--- a/graphblas_algorithms/algorithms/triads.py
+++ b/graphblas_algorithms/algorithms/triads.py
@@ -1,5 +1,4 @@
-from graphblas_algorithms.classes.digraph import DiGraph
-from graphblas_algorithms.classes.graph import Graph
+from graphblas_algorithms import DiGraph, Graph
__all__ = ["is_triad"]
diff --git a/graphblas_algorithms/classes/_utils.py b/graphblas_algorithms/classes/_utils.py
index 92febc5..65ae010 100644
--- a/graphblas_algorithms/classes/_utils.py
+++ b/graphblas_algorithms/classes/_utils.py
@@ -250,3 +250,12 @@ def _cacheit(self, key, func, *args, **kwargs):
if key not in self._cache:
self._cache[key] = func(*args, **kwargs)
return self._cache[key]
+
+
+def renumber_key_to_id(self, indices):
+ """Create `key_to_id` for e.g. a subgraph with node ids from `indices`"""
+ id_to_key = self.id_to_key
+ return {id_to_key[index]: i for i, index in enumerate(indices)}
+ # Alternative (about the same performance)
+ # keys = self.list_to_keys(indices)
+ # return dict(zip(keys, range(len(indices))))
diff --git a/graphblas_algorithms/classes/digraph.py b/graphblas_algorithms/classes/digraph.py
index 83e7356..bae66ae 100644
--- a/graphblas_algorithms/classes/digraph.py
+++ b/graphblas_algorithms/classes/digraph.py
@@ -553,6 +553,7 @@ def __init__(self, incoming_graph_data=None, *, key_to_id=None, **attr):
vector_to_nodeset = _utils.vector_to_nodeset
vector_to_set = _utils.vector_to_set
_cacheit = _utils._cacheit
+ renumber_key_to_id = _utils.renumber_key_to_id
# NetworkX methods
def to_directed_class(self):
@@ -598,6 +599,16 @@ def is_multigraph(self):
def is_directed(self):
return True
+ def to_undirected(self, reciprocal=False, as_view=False, *, name=None):
+ if as_view:
+ raise NotImplementedError("`as_vew=True` is not implemented in `G.to_undirected`")
+ A = self._A
+ if reciprocal:
+ B = binary.any(A & A.T).new(name=name)
+ else:
+ B = binary.any(A | A.T).new(name=name)
+ return Graph(B, key_to_id=self._key_to_id)
+
class MultiDiGraph(DiGraph):
def is_multigraph(self):
diff --git a/graphblas_algorithms/classes/graph.py b/graphblas_algorithms/classes/graph.py
index 03a2893..06f82be 100644
--- a/graphblas_algorithms/classes/graph.py
+++ b/graphblas_algorithms/classes/graph.py
@@ -401,6 +401,7 @@ def __init__(self, incoming_graph_data=None, *, key_to_id=None, **attr):
vector_to_nodeset = _utils.vector_to_nodeset
vector_to_set = _utils.vector_to_set
_cacheit = _utils._cacheit
+ renumber_key_to_id = _utils.renumber_key_to_id
# NetworkX methods
def to_directed_class(self):
diff --git a/graphblas_algorithms/generators/__init__.py b/graphblas_algorithms/generators/__init__.py
new file mode 100644
index 0000000..65a6526
--- /dev/null
+++ b/graphblas_algorithms/generators/__init__.py
@@ -0,0 +1 @@
+from .ego import *
diff --git a/graphblas_algorithms/generators/ego.py b/graphblas_algorithms/generators/ego.py
new file mode 100644
index 0000000..26e9cf9
--- /dev/null
+++ b/graphblas_algorithms/generators/ego.py
@@ -0,0 +1,24 @@
+from ..algorithms.components.connected import _plain_bfs
+from ..algorithms.shortest_paths.weighted import single_source_bellman_ford_path_length
+
+__all__ = ["ego_graph"]
+
+
+def ego_graph(G, n, radius=1, center=True, undirected=False, is_weighted=False):
+ # TODO: should we have an option to keep the output matrix the same size?
+ if undirected and G.is_directed():
+ # NOT COVERED
+ G2 = G.to_undirected()
+ else:
+ G2 = G
+ if is_weighted:
+ v = single_source_bellman_ford_path_length(G2, n, cutoff=radius)
+ else:
+ v = _plain_bfs(G2, n, cutoff=radius)
+ if not center:
+ del v[G._key_to_id[n]]
+
+ indices, _ = v.to_coo(values=False)
+ A = G._A[indices, indices].new(name="ego")
+ key_to_id = G.renumber_key_to_id(indices.tolist())
+ return type(G)(A, key_to_id=key_to_id)
diff --git a/graphblas_algorithms/interface.py b/graphblas_algorithms/interface.py
index 206d19c..94f02a6 100644
--- a/graphblas_algorithms/interface.py
+++ b/graphblas_algorithms/interface.py
@@ -45,6 +45,8 @@ class Dispatcher:
descendants = nxapi.dag.descendants
# Dominating
is_dominating_set = nxapi.dominating.is_dominating_set
+ # Generators
+ ego_graph = nxapi.generators.ego.ego_graph
# Isolate
is_isolate = nxapi.isolate.is_isolate
isolates = nxapi.isolate.isolates
diff --git a/graphblas_algorithms/nxapi/__init__.py b/graphblas_algorithms/nxapi/__init__.py
index 5ddc1fa..2d36017 100644
--- a/graphblas_algorithms/nxapi/__init__.py
+++ b/graphblas_algorithms/nxapi/__init__.py
@@ -7,6 +7,7 @@
from .cuts import *
from .dag import *
from .dominating import *
+from .generators import *
from .isolate import *
from .link_analysis import *
from .operators import *
@@ -23,6 +24,7 @@
from . import cluster
from . import community
from . import components
+from . import generators
from . import link_analysis
from . import operators
from . import shortest_paths
diff --git a/graphblas_algorithms/nxapi/generators/__init__.py b/graphblas_algorithms/nxapi/generators/__init__.py
new file mode 100644
index 0000000..65a6526
--- /dev/null
+++ b/graphblas_algorithms/nxapi/generators/__init__.py
@@ -0,0 +1 @@
+from .ego import *
diff --git a/graphblas_algorithms/nxapi/generators/ego.py b/graphblas_algorithms/nxapi/generators/ego.py
new file mode 100644
index 0000000..e591cb3
--- /dev/null
+++ b/graphblas_algorithms/nxapi/generators/ego.py
@@ -0,0 +1,11 @@
+from graphblas_algorithms import generators
+from graphblas_algorithms.classes.digraph import to_graph
+
+__all__ = ["ego_graph"]
+
+
+def ego_graph(G, n, radius=1, center=True, undirected=False, distance=None):
+ G = to_graph(G, weight=distance)
+ return generators.ego_graph(
+ G, n, radius=radius, center=center, undirected=undirected, is_weighted=distance is not None
+ )
diff --git a/graphblas_algorithms/tests/test_match_nx.py b/graphblas_algorithms/tests/test_match_nx.py
index c50896f..490d1d7 100644
--- a/graphblas_algorithms/tests/test_match_nx.py
+++ b/graphblas_algorithms/tests/test_match_nx.py
@@ -159,3 +159,35 @@ def test_dispatched_funcs_in_nxapi(nx_names_to_info, gb_names_to_info):
print(" ", ":".join(path.rsplit(".", 1)))
if failing: # pragma: no cover
raise AssertionError
+
+
+def test_print_dispatched_not_implemented(nx_names_to_info, gb_names_to_info):
+ """It may be informative to see the results from this to identify functions to implement.
+
+ $ pytest -s -k test_print_dispatched_not_implemented
+ """
+ not_implemented = nx_names_to_info.keys() - gb_names_to_info.keys()
+ fullnames = {next(iter(nx_names_to_info[name])).fullname for name in not_implemented}
+ print()
+ print("=================================================================================")
+ print("Functions dispatched in NetworkX that ARE NOT implemented in graphblas-algorithms")
+ print("---------------------------------------------------------------------------------")
+ for i, name in enumerate(sorted(fullnames)):
+ print(i, name)
+ print("=================================================================================")
+
+
+def test_print_dispatched_implemented(nx_names_to_info, gb_names_to_info):
+ """It may be informative to see the results from this to identify implemented functions.
+
+ $ pytest -s -k test_print_dispatched_implemented
+ """
+ implemented = nx_names_to_info.keys() & gb_names_to_info.keys()
+ fullnames = {next(iter(nx_names_to_info[name])).fullname for name in implemented}
+ print()
+ print("=============================================================================")
+ print("Functions dispatched in NetworkX that ARE implemented in graphblas-algorithms")
+ print("-----------------------------------------------------------------------------")
+ for i, name in enumerate(sorted(fullnames)):
+ print(i, name)
+ print("=============================================================================")
diff --git a/pyproject.toml b/pyproject.toml
index df17600..1772aa2 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -36,7 +36,7 @@ keywords = [
"math",
]
classifiers = [
- "Development Status :: 3 - Alpha",
+ "Development Status :: 4 - Beta",
"License :: OSI Approved :: Apache Software License",
"Operating System :: MacOS :: MacOS X",
"Operating System :: POSIX :: Linux",
@@ -46,6 +46,7 @@ classifiers = [
"Programming Language :: Python :: 3.8",
"Programming Language :: Python :: 3.9",
"Programming Language :: Python :: 3.10",
+ "Programming Language :: Python :: 3.11",
"Programming Language :: Python :: 3 :: Only",
"Intended Audience :: Developers",
"Intended Audience :: Other Audience",
@@ -78,12 +79,8 @@ test = [
"setuptools",
"tomli",
]
-complete = [
- "pytest",
- "networkx >=3.0",
- "scipy >=1.8",
- "setuptools",
- "tomli",
+all = [
+ "graphblas-algorithms[test]",
]
[tool.setuptools]
@@ -104,10 +101,12 @@ packages = [
"graphblas_algorithms.algorithms.tests",
"graphblas_algorithms.algorithms.traversal",
"graphblas_algorithms.classes",
+ "graphblas_algorithms.generators",
"graphblas_algorithms.nxapi",
"graphblas_algorithms.nxapi.centrality",
"graphblas_algorithms.nxapi.community",
"graphblas_algorithms.nxapi.components",
+ "graphblas_algorithms.nxapi.generators",
"graphblas_algorithms.nxapi.link_analysis",
"graphblas_algorithms.nxapi.operators",
"graphblas_algorithms.nxapi.shortest_paths",
### Installation
```
@@ -151,6 +151,8 @@ dispatch pattern shown above.
- descendants
- Dominating
- is_dominating_set
+- Generators
+ - ego_graph
- Isolate
- is_isolate
- isolates
diff --git a/graphblas_algorithms/__init__.py b/graphblas_algorithms/__init__.py
index f9bbcf4..09418f5 100644
--- a/graphblas_algorithms/__init__.py
+++ b/graphblas_algorithms/__init__.py
@@ -3,6 +3,7 @@
from .classes import *
from .algorithms import * # isort:skip
+from .generators import * # isort:skip
try:
__version__ = importlib.metadata.version("graphblas-algorithms")
diff --git a/graphblas_algorithms/algorithms/_bfs.py b/graphblas_algorithms/algorithms/_bfs.py
new file mode 100644
index 0000000..31674ce
--- /dev/null
+++ b/graphblas_algorithms/algorithms/_bfs.py
@@ -0,0 +1,150 @@
+"""BFS routines used by other algorithms"""
+
+import numpy as np
+from graphblas import Matrix, Vector, binary, replace, unary
+from graphblas.semiring import any_pair
+
+
+def _get_cutoff(n, cutoff):
+ if cutoff is None or cutoff >= n:
+ return n # Everything
+ return cutoff + 1 # Inclusive
+
+
+def _plain_bfs(G, source, *, cutoff=None):
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q = Vector(bool, n, name="q")
+ v[index] = True
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for _i in range(1, cutoff):
+ q(~v.S, replace) << any_pair_bool(q @ A)
+ if q.nvals == 0:
+ break
+ v(q.S) << True
+ return v
+
+
+def _bfs_level(G, source, cutoff=None, *, transpose=False, dtype=int):
+ if dtype == bool:
+ dtype = int
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ if transpose and G.is_directed():
+ A = A.T # TODO: should we use "AT" instead?
+ n = A.nrows
+ v = Vector(dtype, n, name="bfs_level")
+ q = Vector(bool, n, name="q")
+ v[index] = 0
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for i in range(1, cutoff):
+ q(~v.S, replace) << any_pair_bool(q @ A)
+ if q.nvals == 0:
+ break
+ v(q.S) << i
+ return v
+
+
+def _bfs_levels(G, nodes, cutoff=None, *, dtype=int):
+ if dtype == bool:
+ dtype = int
+ A = G.get_property("offdiag")
+ n = A.nrows
+ if nodes is None:
+ # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
+ D = Vector(dtype, n, name="bfs_levels_vector")
+ D << 0
+ D = D.diag(name="bfs_levels")
+ else:
+ ids = G.list_to_ids(nodes)
+ D = Matrix.from_coo(
+ np.arange(len(ids), dtype=np.uint64),
+ ids,
+ 0,
+ dtype,
+ nrows=len(ids),
+ ncols=n,
+ name="bfs_levels",
+ )
+ Q = unary.one[bool](D).new(name="Q")
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for i in range(1, cutoff):
+ Q(~D.S, replace) << any_pair_bool(Q @ A)
+ if Q.nvals == 0:
+ break
+ D(Q.S) << i
+ return D
+
+
+# TODO: benchmark this and the version commented out below
+def _plain_bfs_bidirectional(G, source):
+ # Bi-directional BFS w/o symmetrizing the adjacency matrix
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ # XXX: should we use `AT` if available?
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q_out = Vector(bool, n, name="q_out")
+ q_in = Vector(bool, n, name="q_in")
+ v[index] = True
+ q_in[index] = True
+ any_pair_bool = any_pair[bool]
+ is_out_empty = True
+ is_in_empty = False
+ for _i in range(1, n):
+ # Traverse out-edges from the most recent `q_in` and `q_out`
+ if is_out_empty:
+ q_out(~v.S) << any_pair_bool(q_in @ A)
+ else:
+ q_out << binary.any(q_out | q_in)
+ q_out(~v.S, replace) << any_pair_bool(q_out @ A)
+ is_out_empty = q_out.nvals == 0
+ if not is_out_empty:
+ v(q_out.S) << True
+ elif is_in_empty:
+ break
+ # Traverse in-edges from the most recent `q_in` and `q_out`
+ if is_in_empty:
+ q_in(~v.S) << any_pair_bool(A @ q_out)
+ else:
+ q_in << binary.any(q_out | q_in)
+ q_in(~v.S, replace) << any_pair_bool(A @ q_in)
+ is_in_empty = q_in.nvals == 0
+ if not is_in_empty:
+ v(q_in.S) << True
+ elif is_out_empty:
+ break
+ return v
+
+
+"""
+def _plain_bfs_bidirectional(G, source):
+ # Bi-directional BFS w/o symmetrizing the adjacency matrix
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q = Vector(bool, n, name="q")
+ q2 = Vector(bool, n, name="q_2")
+ v[index] = True
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ for _i in range(1, n):
+ q2(~v.S, replace) << any_pair_bool(q @ A)
+ v(q2.S) << True
+ q(~v.S, replace) << any_pair_bool(A @ q)
+ if q.nvals == 0:
+ if q2.nvals == 0:
+ break
+ q, q2 = q2, q
+ elif q2.nvals != 0:
+ q << binary.any(q | q2)
+ return v
+"""
diff --git a/graphblas_algorithms/algorithms/centrality/eigenvector.py b/graphblas_algorithms/algorithms/centrality/eigenvector.py
index 5172f61..e9385f3 100644
--- a/graphblas_algorithms/algorithms/centrality/eigenvector.py
+++ b/graphblas_algorithms/algorithms/centrality/eigenvector.py
@@ -1,11 +1,7 @@
from graphblas import Vector
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import (
- ConvergenceFailure,
- GraphBlasAlgorithmException,
- PointlessConcept,
-)
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure, GraphBlasAlgorithmException, PointlessConcept
__all__ = ["eigenvector_centrality"]
diff --git a/graphblas_algorithms/algorithms/centrality/katz.py b/graphblas_algorithms/algorithms/centrality/katz.py
index 78de982..8087e85 100644
--- a/graphblas_algorithms/algorithms/centrality/katz.py
+++ b/graphblas_algorithms/algorithms/centrality/katz.py
@@ -2,11 +2,8 @@
from graphblas.core.utils import output_type
from graphblas.semiring import plus_first, plus_times
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import (
- ConvergenceFailure,
- GraphBlasAlgorithmException,
-)
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure, GraphBlasAlgorithmException
__all__ = ["katz_centrality"]
diff --git a/graphblas_algorithms/algorithms/components/connected.py b/graphblas_algorithms/algorithms/components/connected.py
index 37c0fc9..fb2f678 100644
--- a/graphblas_algorithms/algorithms/components/connected.py
+++ b/graphblas_algorithms/algorithms/components/connected.py
@@ -1,7 +1,5 @@
-from graphblas import Vector, replace
-from graphblas.semiring import any_pair
-
-from graphblas_algorithms.algorithms.exceptions import PointlessConcept
+from .._bfs import _plain_bfs
+from ..exceptions import PointlessConcept
def is_connected(G):
@@ -12,20 +10,3 @@ def is_connected(G):
def node_connected_component(G, n):
return _plain_bfs(G, n)
-
-
-def _plain_bfs(G, source):
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q = Vector(bool, n, name="q")
- v[index] = True
- q[index] = True
- any_pair_bool = any_pair[bool]
- for _i in range(1, n):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << True
- return v
diff --git a/graphblas_algorithms/algorithms/components/weakly_connected.py b/graphblas_algorithms/algorithms/components/weakly_connected.py
index eb3dc75..99eba78 100644
--- a/graphblas_algorithms/algorithms/components/weakly_connected.py
+++ b/graphblas_algorithms/algorithms/components/weakly_connected.py
@@ -1,77 +1,8 @@
-from graphblas import Vector, binary, replace
-from graphblas.semiring import any_pair
-
-from graphblas_algorithms.algorithms.exceptions import PointlessConcept
+from .._bfs import _plain_bfs_bidirectional
+from ..exceptions import PointlessConcept
def is_weakly_connected(G):
if len(G) == 0:
raise PointlessConcept("Connectivity is undefined for the null graph.")
- return _plain_bfs(G, next(iter(G))).nvals == len(G)
-
-
-# TODO: benchmark this and the version commented out below
-def _plain_bfs(G, source):
- # Bi-directional BFS w/o symmetrizing the adjacency matrix
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- # XXX: should we use `AT` if available?
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q_out = Vector(bool, n, name="q_out")
- q_in = Vector(bool, n, name="q_in")
- v[index] = True
- q_in[index] = True
- any_pair_bool = any_pair[bool]
- is_out_empty = True
- is_in_empty = False
- for _i in range(1, n):
- # Traverse out-edges from the most recent `q_in` and `q_out`
- if is_out_empty:
- q_out(~v.S) << any_pair_bool(q_in @ A)
- else:
- q_out << binary.any(q_out | q_in)
- q_out(~v.S, replace) << any_pair_bool(q_out @ A)
- is_out_empty = q_out.nvals == 0
- if not is_out_empty:
- v(q_out.S) << True
- elif is_in_empty:
- break
- # Traverse in-edges from the most recent `q_in` and `q_out`
- if is_in_empty:
- q_in(~v.S) << any_pair_bool(A @ q_out)
- else:
- q_in << binary.any(q_out | q_in)
- q_in(~v.S, replace) << any_pair_bool(A @ q_in)
- is_in_empty = q_in.nvals == 0
- if not is_in_empty:
- v(q_in.S) << True
- elif is_out_empty:
- break
- return v
-
-
-"""
-def _plain_bfs(G, source):
- # Bi-directional BFS w/o symmetrizing the adjacency matrix
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q = Vector(bool, n, name="q")
- q2 = Vector(bool, n, name="q_2")
- v[index] = True
- q[index] = True
- any_pair_bool = any_pair[bool]
- for _i in range(1, n):
- q2(~v.S, replace) << any_pair_bool(q @ A)
- v(q2.S) << True
- q(~v.S, replace) << any_pair_bool(A @ q)
- if q.nvals == 0:
- if q2.nvals == 0:
- break
- q, q2 = q2, q
- elif q2.nvals != 0:
- q << binary.any(q | q2)
- return v
-"""
+ return _plain_bfs_bidirectional(G, next(iter(G))).nvals == len(G)
diff --git a/graphblas_algorithms/algorithms/core.py b/graphblas_algorithms/algorithms/core.py
index 8133c71..a6ff26d 100644
--- a/graphblas_algorithms/algorithms/core.py
+++ b/graphblas_algorithms/algorithms/core.py
@@ -1,11 +1,12 @@
from graphblas import Matrix, monoid, replace, select, semiring
-from graphblas_algorithms.classes.graph import Graph
+from graphblas_algorithms import Graph
__all__ = ["k_truss"]
def k_truss(G: Graph, k) -> Graph:
+ # TODO: should we have an option to keep the output matrix the same size?
# Ignore self-edges
S = G.get_property("offdiag")
@@ -32,6 +33,5 @@ def k_truss(G: Graph, k) -> Graph:
Ktruss = C[indices, indices].new()
# Convert back to networkx graph with correct node ids
- keys = G.list_to_keys(indices)
- key_to_id = dict(zip(keys, range(len(indices))))
+ key_to_id = G.renumber_key_to_id(indices.tolist())
return Graph(Ktruss, key_to_id=key_to_id)
diff --git a/graphblas_algorithms/algorithms/link_analysis/hits_alg.py b/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
index 515806e..662ac14 100644
--- a/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
+++ b/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
@@ -1,7 +1,7 @@
from graphblas import Vector
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import ConvergenceFailure
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure
__all__ = ["hits"]
diff --git a/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py b/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
index 518c09f..d665e98 100644
--- a/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
+++ b/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
@@ -3,8 +3,9 @@
from graphblas.semiring import plus_first, plus_times
from graphblas_algorithms import Graph
-from graphblas_algorithms.algorithms._helpers import is_converged
-from graphblas_algorithms.algorithms.exceptions import ConvergenceFailure
+
+from .._helpers import is_converged
+from ..exceptions import ConvergenceFailure
__all__ = ["pagerank", "google_matrix"]
diff --git a/graphblas_algorithms/algorithms/shortest_paths/unweighted.py b/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
index 3c8243f..5062e87 100644
--- a/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
+++ b/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
@@ -1,6 +1,6 @@
-import numpy as np
-from graphblas import Matrix, Vector, replace, unary
-from graphblas.semiring import any_pair
+from graphblas import Matrix
+
+from .._bfs import _bfs_level, _bfs_levels
__all__ = [
"single_source_shortest_path_length",
@@ -25,59 +25,3 @@ def all_pairs_shortest_path_length(G, cutoff=None, *, nodes=None, expand_output=
rv[ids, :] = D
return rv
return D
-
-
-def _bfs_level(G, source, cutoff, *, transpose=False):
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- if transpose and G.is_directed():
- A = A.T # TODO: should we use "AT" instead?
- n = A.nrows
- v = Vector(int, n, name="bfs_unweighted")
- q = Vector(bool, n, name="q")
- v[index] = 0
- q[index] = True
- any_pair_bool = any_pair[bool]
- if cutoff is None or cutoff >= n:
- cutoff = n # Everything
- else:
- cutoff += 1 # Inclusive
- for i in range(1, cutoff):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << i
- return v
-
-
-def _bfs_levels(G, nodes, cutoff):
- A = G.get_property("offdiag")
- n = A.nrows
- if nodes is None:
- # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
- D = Vector(int, n, name="bfs_unweighted_vector")
- D << 0
- D = D.diag(name="bfs_unweighted")
- else:
- ids = G.list_to_ids(nodes)
- D = Matrix.from_coo(
- np.arange(len(ids), dtype=np.uint64),
- ids,
- 0,
- int,
- nrows=len(ids),
- ncols=n,
- name="bfs_unweighted",
- )
- Q = unary.one[bool](D).new(name="Q")
- any_pair_bool = any_pair[bool]
- if cutoff is None or cutoff >= n:
- cutoff = n # Everything
- else:
- cutoff += 1 # Inclusive
- for i in range(1, cutoff):
- Q(~D.S, replace) << any_pair_bool(Q @ A)
- if Q.nvals == 0:
- break
- D(Q.S) << i
- return D
diff --git a/graphblas_algorithms/algorithms/shortest_paths/weighted.py b/graphblas_algorithms/algorithms/shortest_paths/weighted.py
index 8e6efef..fddf672 100644
--- a/graphblas_algorithms/algorithms/shortest_paths/weighted.py
+++ b/graphblas_algorithms/algorithms/shortest_paths/weighted.py
@@ -2,6 +2,7 @@
from graphblas import Matrix, Vector, binary, monoid, replace, select, unary
from graphblas.semiring import any_pair, min_plus
+from .._bfs import _bfs_level, _bfs_levels
from ..exceptions import Unbounded
__all__ = [
@@ -11,14 +12,16 @@
]
-def single_source_bellman_ford_path_length(G, source):
+def single_source_bellman_ford_path_length(G, source, *, cutoff=None):
# No need for `is_weighted=` keyword, b/c this is assumed to be weighted (I think)
index = G._key_to_id[source]
if G.get_property("is_iso"):
# If the edges are iso-valued (and positive), then we can simply do level BFS
is_negative, iso_value = G.get_properties("has_negative_edges+ iso_value")
if not is_negative:
- d = _bfs_level(G, source, dtype=iso_value.dtype)
+ if cutoff is not None:
+ cutoff = int(cutoff // iso_value)
+ d = _bfs_level(G, source, cutoff, dtype=iso_value.dtype)
if iso_value != 1:
d *= iso_value
return d
@@ -49,6 +52,8 @@ def single_source_bellman_ford_path_length(G, source):
# `cur` is the current frontier of values that improved in the previous iteration.
# This means that in this iteration we drop values from `cur` that are not better.
cur << min_plus(cur @ A)
+ if cutoff is not None:
+ cur << select.valuele(cur, cutoff)
# Mask is True where cur not in d or cur < d
mask << one(cur)
@@ -63,6 +68,8 @@ def single_source_bellman_ford_path_length(G, source):
else:
# Check for negative cycle when for loop completes without breaking
cur << min_plus(cur @ A)
+ if cutoff is not None:
+ cur << select.valuele(cur, cutoff)
mask << binary.lt(cur & d)
if mask.reduce(monoid.lor):
raise Unbounded("Negative cycle detected.")
@@ -157,56 +164,6 @@ def bellman_ford_path_lengths(G, nodes=None, *, expand_output=False):
return D
-def _bfs_level(G, source, *, dtype=int):
- if dtype == bool:
- dtype = int
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(dtype, n, name="bfs_level")
- q = Vector(bool, n, name="q")
- v[index] = 0
- q[index] = True
- any_pair_bool = any_pair[bool]
- for i in range(1, n):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << i
- return v
-
-
-def _bfs_levels(G, nodes=None, *, dtype=int):
- if dtype == bool:
- dtype = int
- A = G.get_property("offdiag")
- n = A.nrows
- if nodes is None:
- # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
- D = Vector(dtype, n, name="bfs_levels_vector")
- D << 0
- D = D.diag(name="bfs_levels")
- else:
- ids = G.list_to_ids(nodes)
- D = Matrix.from_coo(
- np.arange(len(ids), dtype=np.uint64),
- ids,
- 0,
- dtype,
- nrows=len(ids),
- ncols=n,
- name="bfs_levels",
- )
- Q = unary.one[bool](D).new(name="Q")
- any_pair_bool = any_pair[bool]
- for i in range(1, n):
- Q(~D.S, replace) << any_pair_bool(Q @ A)
- if Q.nvals == 0:
- break
- D(Q.S) << i
- return D
-
-
def negative_edge_cycle(G):
# TODO: use a heuristic to try to stop early
if G.is_directed():
diff --git a/graphblas_algorithms/algorithms/triads.py b/graphblas_algorithms/algorithms/triads.py
index 54702c7..e6ec2be 100644
--- a/graphblas_algorithms/algorithms/triads.py
+++ b/graphblas_algorithms/algorithms/triads.py
@@ -1,5 +1,4 @@
-from graphblas_algorithms.classes.digraph import DiGraph
-from graphblas_algorithms.classes.graph import Graph
+from graphblas_algorithms import DiGraph, Graph
__all__ = ["is_triad"]
diff --git a/graphblas_algorithms/classes/_utils.py b/graphblas_algorithms/classes/_utils.py
index 92febc5..65ae010 100644
--- a/graphblas_algorithms/classes/_utils.py
+++ b/graphblas_algorithms/classes/_utils.py
@@ -250,3 +250,12 @@ def _cacheit(self, key, func, *args, **kwargs):
if key not in self._cache:
self._cache[key] = func(*args, **kwargs)
return self._cache[key]
+
+
+def renumber_key_to_id(self, indices):
+ """Create `key_to_id` for e.g. a subgraph with node ids from `indices`"""
+ id_to_key = self.id_to_key
+ return {id_to_key[index]: i for i, index in enumerate(indices)}
+ # Alternative (about the same performance)
+ # keys = self.list_to_keys(indices)
+ # return dict(zip(keys, range(len(indices))))
diff --git a/graphblas_algorithms/classes/digraph.py b/graphblas_algorithms/classes/digraph.py
index 83e7356..bae66ae 100644
--- a/graphblas_algorithms/classes/digraph.py
+++ b/graphblas_algorithms/classes/digraph.py
@@ -553,6 +553,7 @@ def __init__(self, incoming_graph_data=None, *, key_to_id=None, **attr):
vector_to_nodeset = _utils.vector_to_nodeset
vector_to_set = _utils.vector_to_set
_cacheit = _utils._cacheit
+ renumber_key_to_id = _utils.renumber_key_to_id
# NetworkX methods
def to_directed_class(self):
@@ -598,6 +599,16 @@ def is_multigraph(self):
def is_directed(self):
return True
+ def to_undirected(self, reciprocal=False, as_view=False, *, name=None):
+ if as_view:
+ raise NotImplementedError("`as_vew=True` is not implemented in `G.to_undirected`")
+ A = self._A
+ if reciprocal:
+ B = binary.any(A & A.T).new(name=name)
+ else:
+ B = binary.any(A | A.T).new(name=name)
+ return Graph(B, key_to_id=self._key_to_id)
+
class MultiDiGraph(DiGraph):
def is_multigraph(self):
diff --git a/graphblas_algorithms/classes/graph.py b/graphblas_algorithms/classes/graph.py
index 03a2893..06f82be 100644
--- a/graphblas_algorithms/classes/graph.py
+++ b/graphblas_algorithms/classes/graph.py
@@ -401,6 +401,7 @@ def __init__(self, incoming_graph_data=None, *, key_to_id=None, **attr):
vector_to_nodeset = _utils.vector_to_nodeset
vector_to_set = _utils.vector_to_set
_cacheit = _utils._cacheit
+ renumber_key_to_id = _utils.renumber_key_to_id
# NetworkX methods
def to_directed_class(self):
diff --git a/graphblas_algorithms/generators/__init__.py b/graphblas_algorithms/generators/__init__.py
new file mode 100644
index 0000000..65a6526
--- /dev/null
+++ b/graphblas_algorithms/generators/__init__.py
@@ -0,0 +1 @@
+from .ego import *
diff --git a/graphblas_algorithms/generators/ego.py b/graphblas_algorithms/generators/ego.py
new file mode 100644
index 0000000..26e9cf9
--- /dev/null
+++ b/graphblas_algorithms/generators/ego.py
@@ -0,0 +1,24 @@
+from ..algorithms.components.connected import _plain_bfs
+from ..algorithms.shortest_paths.weighted import single_source_bellman_ford_path_length
+
+__all__ = ["ego_graph"]
+
+
+def ego_graph(G, n, radius=1, center=True, undirected=False, is_weighted=False):
+ # TODO: should we have an option to keep the output matrix the same size?
+ if undirected and G.is_directed():
+ # NOT COVERED
+ G2 = G.to_undirected()
+ else:
+ G2 = G
+ if is_weighted:
+ v = single_source_bellman_ford_path_length(G2, n, cutoff=radius)
+ else:
+ v = _plain_bfs(G2, n, cutoff=radius)
+ if not center:
+ del v[G._key_to_id[n]]
+
+ indices, _ = v.to_coo(values=False)
+ A = G._A[indices, indices].new(name="ego")
+ key_to_id = G.renumber_key_to_id(indices.tolist())
+ return type(G)(A, key_to_id=key_to_id)
diff --git a/graphblas_algorithms/interface.py b/graphblas_algorithms/interface.py
index 206d19c..94f02a6 100644
--- a/graphblas_algorithms/interface.py
+++ b/graphblas_algorithms/interface.py
@@ -45,6 +45,8 @@ class Dispatcher:
descendants = nxapi.dag.descendants
# Dominating
is_dominating_set = nxapi.dominating.is_dominating_set
+ # Generators
+ ego_graph = nxapi.generators.ego.ego_graph
# Isolate
is_isolate = nxapi.isolate.is_isolate
isolates = nxapi.isolate.isolates
diff --git a/graphblas_algorithms/nxapi/__init__.py b/graphblas_algorithms/nxapi/__init__.py
index 5ddc1fa..2d36017 100644
--- a/graphblas_algorithms/nxapi/__init__.py
+++ b/graphblas_algorithms/nxapi/__init__.py
@@ -7,6 +7,7 @@
from .cuts import *
from .dag import *
from .dominating import *
+from .generators import *
from .isolate import *
from .link_analysis import *
from .operators import *
@@ -23,6 +24,7 @@
from . import cluster
from . import community
from . import components
+from . import generators
from . import link_analysis
from . import operators
from . import shortest_paths
diff --git a/graphblas_algorithms/nxapi/generators/__init__.py b/graphblas_algorithms/nxapi/generators/__init__.py
new file mode 100644
index 0000000..65a6526
--- /dev/null
+++ b/graphblas_algorithms/nxapi/generators/__init__.py
@@ -0,0 +1 @@
+from .ego import *
diff --git a/graphblas_algorithms/nxapi/generators/ego.py b/graphblas_algorithms/nxapi/generators/ego.py
new file mode 100644
index 0000000..e591cb3
--- /dev/null
+++ b/graphblas_algorithms/nxapi/generators/ego.py
@@ -0,0 +1,11 @@
+from graphblas_algorithms import generators
+from graphblas_algorithms.classes.digraph import to_graph
+
+__all__ = ["ego_graph"]
+
+
+def ego_graph(G, n, radius=1, center=True, undirected=False, distance=None):
+ G = to_graph(G, weight=distance)
+ return generators.ego_graph(
+ G, n, radius=radius, center=center, undirected=undirected, is_weighted=distance is not None
+ )
diff --git a/graphblas_algorithms/tests/test_match_nx.py b/graphblas_algorithms/tests/test_match_nx.py
index c50896f..490d1d7 100644
--- a/graphblas_algorithms/tests/test_match_nx.py
+++ b/graphblas_algorithms/tests/test_match_nx.py
@@ -159,3 +159,35 @@ def test_dispatched_funcs_in_nxapi(nx_names_to_info, gb_names_to_info):
print(" ", ":".join(path.rsplit(".", 1)))
if failing: # pragma: no cover
raise AssertionError
+
+
+def test_print_dispatched_not_implemented(nx_names_to_info, gb_names_to_info):
+ """It may be informative to see the results from this to identify functions to implement.
+
+ $ pytest -s -k test_print_dispatched_not_implemented
+ """
+ not_implemented = nx_names_to_info.keys() - gb_names_to_info.keys()
+ fullnames = {next(iter(nx_names_to_info[name])).fullname for name in not_implemented}
+ print()
+ print("=================================================================================")
+ print("Functions dispatched in NetworkX that ARE NOT implemented in graphblas-algorithms")
+ print("---------------------------------------------------------------------------------")
+ for i, name in enumerate(sorted(fullnames)):
+ print(i, name)
+ print("=================================================================================")
+
+
+def test_print_dispatched_implemented(nx_names_to_info, gb_names_to_info):
+ """It may be informative to see the results from this to identify implemented functions.
+
+ $ pytest -s -k test_print_dispatched_implemented
+ """
+ implemented = nx_names_to_info.keys() & gb_names_to_info.keys()
+ fullnames = {next(iter(nx_names_to_info[name])).fullname for name in implemented}
+ print()
+ print("=============================================================================")
+ print("Functions dispatched in NetworkX that ARE implemented in graphblas-algorithms")
+ print("-----------------------------------------------------------------------------")
+ for i, name in enumerate(sorted(fullnames)):
+ print(i, name)
+ print("=============================================================================")
diff --git a/pyproject.toml b/pyproject.toml
index df17600..1772aa2 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -36,7 +36,7 @@ keywords = [
"math",
]
classifiers = [
- "Development Status :: 3 - Alpha",
+ "Development Status :: 4 - Beta",
"License :: OSI Approved :: Apache Software License",
"Operating System :: MacOS :: MacOS X",
"Operating System :: POSIX :: Linux",
@@ -46,6 +46,7 @@ classifiers = [
"Programming Language :: Python :: 3.8",
"Programming Language :: Python :: 3.9",
"Programming Language :: Python :: 3.10",
+ "Programming Language :: Python :: 3.11",
"Programming Language :: Python :: 3 :: Only",
"Intended Audience :: Developers",
"Intended Audience :: Other Audience",
@@ -78,12 +79,8 @@ test = [
"setuptools",
"tomli",
]
-complete = [
- "pytest",
- "networkx >=3.0",
- "scipy >=1.8",
- "setuptools",
- "tomli",
+all = [
+ "graphblas-algorithms[test]",
]
[tool.setuptools]
@@ -104,10 +101,12 @@ packages = [
"graphblas_algorithms.algorithms.tests",
"graphblas_algorithms.algorithms.traversal",
"graphblas_algorithms.classes",
+ "graphblas_algorithms.generators",
"graphblas_algorithms.nxapi",
"graphblas_algorithms.nxapi.centrality",
"graphblas_algorithms.nxapi.community",
"graphblas_algorithms.nxapi.components",
+ "graphblas_algorithms.nxapi.generators",
"graphblas_algorithms.nxapi.link_analysis",
"graphblas_algorithms.nxapi.operators",
"graphblas_algorithms.nxapi.shortest_paths",
 +
+
 +
+ ### Installation
```
@@ -151,6 +151,8 @@ dispatch pattern shown above.
- descendants
- Dominating
- is_dominating_set
+- Generators
+ - ego_graph
- Isolate
- is_isolate
- isolates
diff --git a/graphblas_algorithms/__init__.py b/graphblas_algorithms/__init__.py
index f9bbcf4..09418f5 100644
--- a/graphblas_algorithms/__init__.py
+++ b/graphblas_algorithms/__init__.py
@@ -3,6 +3,7 @@
from .classes import *
from .algorithms import * # isort:skip
+from .generators import * # isort:skip
try:
__version__ = importlib.metadata.version("graphblas-algorithms")
diff --git a/graphblas_algorithms/algorithms/_bfs.py b/graphblas_algorithms/algorithms/_bfs.py
new file mode 100644
index 0000000..31674ce
--- /dev/null
+++ b/graphblas_algorithms/algorithms/_bfs.py
@@ -0,0 +1,150 @@
+"""BFS routines used by other algorithms"""
+
+import numpy as np
+from graphblas import Matrix, Vector, binary, replace, unary
+from graphblas.semiring import any_pair
+
+
+def _get_cutoff(n, cutoff):
+ if cutoff is None or cutoff >= n:
+ return n # Everything
+ return cutoff + 1 # Inclusive
+
+
+def _plain_bfs(G, source, *, cutoff=None):
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q = Vector(bool, n, name="q")
+ v[index] = True
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for _i in range(1, cutoff):
+ q(~v.S, replace) << any_pair_bool(q @ A)
+ if q.nvals == 0:
+ break
+ v(q.S) << True
+ return v
+
+
+def _bfs_level(G, source, cutoff=None, *, transpose=False, dtype=int):
+ if dtype == bool:
+ dtype = int
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ if transpose and G.is_directed():
+ A = A.T # TODO: should we use "AT" instead?
+ n = A.nrows
+ v = Vector(dtype, n, name="bfs_level")
+ q = Vector(bool, n, name="q")
+ v[index] = 0
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for i in range(1, cutoff):
+ q(~v.S, replace) << any_pair_bool(q @ A)
+ if q.nvals == 0:
+ break
+ v(q.S) << i
+ return v
+
+
+def _bfs_levels(G, nodes, cutoff=None, *, dtype=int):
+ if dtype == bool:
+ dtype = int
+ A = G.get_property("offdiag")
+ n = A.nrows
+ if nodes is None:
+ # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
+ D = Vector(dtype, n, name="bfs_levels_vector")
+ D << 0
+ D = D.diag(name="bfs_levels")
+ else:
+ ids = G.list_to_ids(nodes)
+ D = Matrix.from_coo(
+ np.arange(len(ids), dtype=np.uint64),
+ ids,
+ 0,
+ dtype,
+ nrows=len(ids),
+ ncols=n,
+ name="bfs_levels",
+ )
+ Q = unary.one[bool](D).new(name="Q")
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for i in range(1, cutoff):
+ Q(~D.S, replace) << any_pair_bool(Q @ A)
+ if Q.nvals == 0:
+ break
+ D(Q.S) << i
+ return D
+
+
+# TODO: benchmark this and the version commented out below
+def _plain_bfs_bidirectional(G, source):
+ # Bi-directional BFS w/o symmetrizing the adjacency matrix
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ # XXX: should we use `AT` if available?
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q_out = Vector(bool, n, name="q_out")
+ q_in = Vector(bool, n, name="q_in")
+ v[index] = True
+ q_in[index] = True
+ any_pair_bool = any_pair[bool]
+ is_out_empty = True
+ is_in_empty = False
+ for _i in range(1, n):
+ # Traverse out-edges from the most recent `q_in` and `q_out`
+ if is_out_empty:
+ q_out(~v.S) << any_pair_bool(q_in @ A)
+ else:
+ q_out << binary.any(q_out | q_in)
+ q_out(~v.S, replace) << any_pair_bool(q_out @ A)
+ is_out_empty = q_out.nvals == 0
+ if not is_out_empty:
+ v(q_out.S) << True
+ elif is_in_empty:
+ break
+ # Traverse in-edges from the most recent `q_in` and `q_out`
+ if is_in_empty:
+ q_in(~v.S) << any_pair_bool(A @ q_out)
+ else:
+ q_in << binary.any(q_out | q_in)
+ q_in(~v.S, replace) << any_pair_bool(A @ q_in)
+ is_in_empty = q_in.nvals == 0
+ if not is_in_empty:
+ v(q_in.S) << True
+ elif is_out_empty:
+ break
+ return v
+
+
+"""
+def _plain_bfs_bidirectional(G, source):
+ # Bi-directional BFS w/o symmetrizing the adjacency matrix
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q = Vector(bool, n, name="q")
+ q2 = Vector(bool, n, name="q_2")
+ v[index] = True
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ for _i in range(1, n):
+ q2(~v.S, replace) << any_pair_bool(q @ A)
+ v(q2.S) << True
+ q(~v.S, replace) << any_pair_bool(A @ q)
+ if q.nvals == 0:
+ if q2.nvals == 0:
+ break
+ q, q2 = q2, q
+ elif q2.nvals != 0:
+ q << binary.any(q | q2)
+ return v
+"""
diff --git a/graphblas_algorithms/algorithms/centrality/eigenvector.py b/graphblas_algorithms/algorithms/centrality/eigenvector.py
index 5172f61..e9385f3 100644
--- a/graphblas_algorithms/algorithms/centrality/eigenvector.py
+++ b/graphblas_algorithms/algorithms/centrality/eigenvector.py
@@ -1,11 +1,7 @@
from graphblas import Vector
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import (
- ConvergenceFailure,
- GraphBlasAlgorithmException,
- PointlessConcept,
-)
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure, GraphBlasAlgorithmException, PointlessConcept
__all__ = ["eigenvector_centrality"]
diff --git a/graphblas_algorithms/algorithms/centrality/katz.py b/graphblas_algorithms/algorithms/centrality/katz.py
index 78de982..8087e85 100644
--- a/graphblas_algorithms/algorithms/centrality/katz.py
+++ b/graphblas_algorithms/algorithms/centrality/katz.py
@@ -2,11 +2,8 @@
from graphblas.core.utils import output_type
from graphblas.semiring import plus_first, plus_times
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import (
- ConvergenceFailure,
- GraphBlasAlgorithmException,
-)
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure, GraphBlasAlgorithmException
__all__ = ["katz_centrality"]
diff --git a/graphblas_algorithms/algorithms/components/connected.py b/graphblas_algorithms/algorithms/components/connected.py
index 37c0fc9..fb2f678 100644
--- a/graphblas_algorithms/algorithms/components/connected.py
+++ b/graphblas_algorithms/algorithms/components/connected.py
@@ -1,7 +1,5 @@
-from graphblas import Vector, replace
-from graphblas.semiring import any_pair
-
-from graphblas_algorithms.algorithms.exceptions import PointlessConcept
+from .._bfs import _plain_bfs
+from ..exceptions import PointlessConcept
def is_connected(G):
@@ -12,20 +10,3 @@ def is_connected(G):
def node_connected_component(G, n):
return _plain_bfs(G, n)
-
-
-def _plain_bfs(G, source):
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q = Vector(bool, n, name="q")
- v[index] = True
- q[index] = True
- any_pair_bool = any_pair[bool]
- for _i in range(1, n):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << True
- return v
diff --git a/graphblas_algorithms/algorithms/components/weakly_connected.py b/graphblas_algorithms/algorithms/components/weakly_connected.py
index eb3dc75..99eba78 100644
--- a/graphblas_algorithms/algorithms/components/weakly_connected.py
+++ b/graphblas_algorithms/algorithms/components/weakly_connected.py
@@ -1,77 +1,8 @@
-from graphblas import Vector, binary, replace
-from graphblas.semiring import any_pair
-
-from graphblas_algorithms.algorithms.exceptions import PointlessConcept
+from .._bfs import _plain_bfs_bidirectional
+from ..exceptions import PointlessConcept
def is_weakly_connected(G):
if len(G) == 0:
raise PointlessConcept("Connectivity is undefined for the null graph.")
- return _plain_bfs(G, next(iter(G))).nvals == len(G)
-
-
-# TODO: benchmark this and the version commented out below
-def _plain_bfs(G, source):
- # Bi-directional BFS w/o symmetrizing the adjacency matrix
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- # XXX: should we use `AT` if available?
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q_out = Vector(bool, n, name="q_out")
- q_in = Vector(bool, n, name="q_in")
- v[index] = True
- q_in[index] = True
- any_pair_bool = any_pair[bool]
- is_out_empty = True
- is_in_empty = False
- for _i in range(1, n):
- # Traverse out-edges from the most recent `q_in` and `q_out`
- if is_out_empty:
- q_out(~v.S) << any_pair_bool(q_in @ A)
- else:
- q_out << binary.any(q_out | q_in)
- q_out(~v.S, replace) << any_pair_bool(q_out @ A)
- is_out_empty = q_out.nvals == 0
- if not is_out_empty:
- v(q_out.S) << True
- elif is_in_empty:
- break
- # Traverse in-edges from the most recent `q_in` and `q_out`
- if is_in_empty:
- q_in(~v.S) << any_pair_bool(A @ q_out)
- else:
- q_in << binary.any(q_out | q_in)
- q_in(~v.S, replace) << any_pair_bool(A @ q_in)
- is_in_empty = q_in.nvals == 0
- if not is_in_empty:
- v(q_in.S) << True
- elif is_out_empty:
- break
- return v
-
-
-"""
-def _plain_bfs(G, source):
- # Bi-directional BFS w/o symmetrizing the adjacency matrix
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q = Vector(bool, n, name="q")
- q2 = Vector(bool, n, name="q_2")
- v[index] = True
- q[index] = True
- any_pair_bool = any_pair[bool]
- for _i in range(1, n):
- q2(~v.S, replace) << any_pair_bool(q @ A)
- v(q2.S) << True
- q(~v.S, replace) << any_pair_bool(A @ q)
- if q.nvals == 0:
- if q2.nvals == 0:
- break
- q, q2 = q2, q
- elif q2.nvals != 0:
- q << binary.any(q | q2)
- return v
-"""
+ return _plain_bfs_bidirectional(G, next(iter(G))).nvals == len(G)
diff --git a/graphblas_algorithms/algorithms/core.py b/graphblas_algorithms/algorithms/core.py
index 8133c71..a6ff26d 100644
--- a/graphblas_algorithms/algorithms/core.py
+++ b/graphblas_algorithms/algorithms/core.py
@@ -1,11 +1,12 @@
from graphblas import Matrix, monoid, replace, select, semiring
-from graphblas_algorithms.classes.graph import Graph
+from graphblas_algorithms import Graph
__all__ = ["k_truss"]
def k_truss(G: Graph, k) -> Graph:
+ # TODO: should we have an option to keep the output matrix the same size?
# Ignore self-edges
S = G.get_property("offdiag")
@@ -32,6 +33,5 @@ def k_truss(G: Graph, k) -> Graph:
Ktruss = C[indices, indices].new()
# Convert back to networkx graph with correct node ids
- keys = G.list_to_keys(indices)
- key_to_id = dict(zip(keys, range(len(indices))))
+ key_to_id = G.renumber_key_to_id(indices.tolist())
return Graph(Ktruss, key_to_id=key_to_id)
diff --git a/graphblas_algorithms/algorithms/link_analysis/hits_alg.py b/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
index 515806e..662ac14 100644
--- a/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
+++ b/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
@@ -1,7 +1,7 @@
from graphblas import Vector
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import ConvergenceFailure
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure
__all__ = ["hits"]
diff --git a/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py b/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
index 518c09f..d665e98 100644
--- a/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
+++ b/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
@@ -3,8 +3,9 @@
from graphblas.semiring import plus_first, plus_times
from graphblas_algorithms import Graph
-from graphblas_algorithms.algorithms._helpers import is_converged
-from graphblas_algorithms.algorithms.exceptions import ConvergenceFailure
+
+from .._helpers import is_converged
+from ..exceptions import ConvergenceFailure
__all__ = ["pagerank", "google_matrix"]
diff --git a/graphblas_algorithms/algorithms/shortest_paths/unweighted.py b/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
index 3c8243f..5062e87 100644
--- a/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
+++ b/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
@@ -1,6 +1,6 @@
-import numpy as np
-from graphblas import Matrix, Vector, replace, unary
-from graphblas.semiring import any_pair
+from graphblas import Matrix
+
+from .._bfs import _bfs_level, _bfs_levels
__all__ = [
"single_source_shortest_path_length",
@@ -25,59 +25,3 @@ def all_pairs_shortest_path_length(G, cutoff=None, *, nodes=None, expand_output=
rv[ids, :] = D
return rv
return D
-
-
-def _bfs_level(G, source, cutoff, *, transpose=False):
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- if transpose and G.is_directed():
- A = A.T # TODO: should we use "AT" instead?
- n = A.nrows
- v = Vector(int, n, name="bfs_unweighted")
- q = Vector(bool, n, name="q")
- v[index] = 0
- q[index] = True
- any_pair_bool = any_pair[bool]
- if cutoff is None or cutoff >= n:
- cutoff = n # Everything
- else:
- cutoff += 1 # Inclusive
- for i in range(1, cutoff):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << i
- return v
-
-
-def _bfs_levels(G, nodes, cutoff):
- A = G.get_property("offdiag")
- n = A.nrows
- if nodes is None:
- # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
- D = Vector(int, n, name="bfs_unweighted_vector")
- D << 0
- D = D.diag(name="bfs_unweighted")
- else:
- ids = G.list_to_ids(nodes)
- D = Matrix.from_coo(
- np.arange(len(ids), dtype=np.uint64),
- ids,
- 0,
- int,
- nrows=len(ids),
- ncols=n,
- name="bfs_unweighted",
- )
- Q = unary.one[bool](D).new(name="Q")
- any_pair_bool = any_pair[bool]
- if cutoff is None or cutoff >= n:
- cutoff = n # Everything
- else:
- cutoff += 1 # Inclusive
- for i in range(1, cutoff):
- Q(~D.S, replace) << any_pair_bool(Q @ A)
- if Q.nvals == 0:
- break
- D(Q.S) << i
- return D
diff --git a/graphblas_algorithms/algorithms/shortest_paths/weighted.py b/graphblas_algorithms/algorithms/shortest_paths/weighted.py
index 8e6efef..fddf672 100644
--- a/graphblas_algorithms/algorithms/shortest_paths/weighted.py
+++ b/graphblas_algorithms/algorithms/shortest_paths/weighted.py
@@ -2,6 +2,7 @@
from graphblas import Matrix, Vector, binary, monoid, replace, select, unary
from graphblas.semiring import any_pair, min_plus
+from .._bfs import _bfs_level, _bfs_levels
from ..exceptions import Unbounded
__all__ = [
@@ -11,14 +12,16 @@
]
-def single_source_bellman_ford_path_length(G, source):
+def single_source_bellman_ford_path_length(G, source, *, cutoff=None):
# No need for `is_weighted=` keyword, b/c this is assumed to be weighted (I think)
index = G._key_to_id[source]
if G.get_property("is_iso"):
# If the edges are iso-valued (and positive), then we can simply do level BFS
is_negative, iso_value = G.get_properties("has_negative_edges+ iso_value")
if not is_negative:
- d = _bfs_level(G, source, dtype=iso_value.dtype)
+ if cutoff is not None:
+ cutoff = int(cutoff // iso_value)
+ d = _bfs_level(G, source, cutoff, dtype=iso_value.dtype)
if iso_value != 1:
d *= iso_value
return d
@@ -49,6 +52,8 @@ def single_source_bellman_ford_path_length(G, source):
# `cur` is the current frontier of values that improved in the previous iteration.
# This means that in this iteration we drop values from `cur` that are not better.
cur << min_plus(cur @ A)
+ if cutoff is not None:
+ cur << select.valuele(cur, cutoff)
# Mask is True where cur not in d or cur < d
mask << one(cur)
@@ -63,6 +68,8 @@ def single_source_bellman_ford_path_length(G, source):
else:
# Check for negative cycle when for loop completes without breaking
cur << min_plus(cur @ A)
+ if cutoff is not None:
+ cur << select.valuele(cur, cutoff)
mask << binary.lt(cur & d)
if mask.reduce(monoid.lor):
raise Unbounded("Negative cycle detected.")
@@ -157,56 +164,6 @@ def bellman_ford_path_lengths(G, nodes=None, *, expand_output=False):
return D
-def _bfs_level(G, source, *, dtype=int):
- if dtype == bool:
- dtype = int
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(dtype, n, name="bfs_level")
- q = Vector(bool, n, name="q")
- v[index] = 0
- q[index] = True
- any_pair_bool = any_pair[bool]
- for i in range(1, n):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << i
- return v
-
-
-def _bfs_levels(G, nodes=None, *, dtype=int):
- if dtype == bool:
- dtype = int
- A = G.get_property("offdiag")
- n = A.nrows
- if nodes is None:
- # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
- D = Vector(dtype, n, name="bfs_levels_vector")
- D << 0
- D = D.diag(name="bfs_levels")
- else:
- ids = G.list_to_ids(nodes)
- D = Matrix.from_coo(
- np.arange(len(ids), dtype=np.uint64),
- ids,
- 0,
- dtype,
- nrows=len(ids),
- ncols=n,
- name="bfs_levels",
- )
- Q = unary.one[bool](D).new(name="Q")
- any_pair_bool = any_pair[bool]
- for i in range(1, n):
- Q(~D.S, replace) << any_pair_bool(Q @ A)
- if Q.nvals == 0:
- break
- D(Q.S) << i
- return D
-
-
def negative_edge_cycle(G):
# TODO: use a heuristic to try to stop early
if G.is_directed():
diff --git a/graphblas_algorithms/algorithms/triads.py b/graphblas_algorithms/algorithms/triads.py
index 54702c7..e6ec2be 100644
--- a/graphblas_algorithms/algorithms/triads.py
+++ b/graphblas_algorithms/algorithms/triads.py
@@ -1,5 +1,4 @@
-from graphblas_algorithms.classes.digraph import DiGraph
-from graphblas_algorithms.classes.graph import Graph
+from graphblas_algorithms import DiGraph, Graph
__all__ = ["is_triad"]
diff --git a/graphblas_algorithms/classes/_utils.py b/graphblas_algorithms/classes/_utils.py
index 92febc5..65ae010 100644
--- a/graphblas_algorithms/classes/_utils.py
+++ b/graphblas_algorithms/classes/_utils.py
@@ -250,3 +250,12 @@ def _cacheit(self, key, func, *args, **kwargs):
if key not in self._cache:
self._cache[key] = func(*args, **kwargs)
return self._cache[key]
+
+
+def renumber_key_to_id(self, indices):
+ """Create `key_to_id` for e.g. a subgraph with node ids from `indices`"""
+ id_to_key = self.id_to_key
+ return {id_to_key[index]: i for i, index in enumerate(indices)}
+ # Alternative (about the same performance)
+ # keys = self.list_to_keys(indices)
+ # return dict(zip(keys, range(len(indices))))
diff --git a/graphblas_algorithms/classes/digraph.py b/graphblas_algorithms/classes/digraph.py
index 83e7356..bae66ae 100644
--- a/graphblas_algorithms/classes/digraph.py
+++ b/graphblas_algorithms/classes/digraph.py
@@ -553,6 +553,7 @@ def __init__(self, incoming_graph_data=None, *, key_to_id=None, **attr):
vector_to_nodeset = _utils.vector_to_nodeset
vector_to_set = _utils.vector_to_set
_cacheit = _utils._cacheit
+ renumber_key_to_id = _utils.renumber_key_to_id
# NetworkX methods
def to_directed_class(self):
@@ -598,6 +599,16 @@ def is_multigraph(self):
def is_directed(self):
return True
+ def to_undirected(self, reciprocal=False, as_view=False, *, name=None):
+ if as_view:
+ raise NotImplementedError("`as_vew=True` is not implemented in `G.to_undirected`")
+ A = self._A
+ if reciprocal:
+ B = binary.any(A & A.T).new(name=name)
+ else:
+ B = binary.any(A | A.T).new(name=name)
+ return Graph(B, key_to_id=self._key_to_id)
+
class MultiDiGraph(DiGraph):
def is_multigraph(self):
diff --git a/graphblas_algorithms/classes/graph.py b/graphblas_algorithms/classes/graph.py
index 03a2893..06f82be 100644
--- a/graphblas_algorithms/classes/graph.py
+++ b/graphblas_algorithms/classes/graph.py
@@ -401,6 +401,7 @@ def __init__(self, incoming_graph_data=None, *, key_to_id=None, **attr):
vector_to_nodeset = _utils.vector_to_nodeset
vector_to_set = _utils.vector_to_set
_cacheit = _utils._cacheit
+ renumber_key_to_id = _utils.renumber_key_to_id
# NetworkX methods
def to_directed_class(self):
diff --git a/graphblas_algorithms/generators/__init__.py b/graphblas_algorithms/generators/__init__.py
new file mode 100644
index 0000000..65a6526
--- /dev/null
+++ b/graphblas_algorithms/generators/__init__.py
@@ -0,0 +1 @@
+from .ego import *
diff --git a/graphblas_algorithms/generators/ego.py b/graphblas_algorithms/generators/ego.py
new file mode 100644
index 0000000..26e9cf9
--- /dev/null
+++ b/graphblas_algorithms/generators/ego.py
@@ -0,0 +1,24 @@
+from ..algorithms.components.connected import _plain_bfs
+from ..algorithms.shortest_paths.weighted import single_source_bellman_ford_path_length
+
+__all__ = ["ego_graph"]
+
+
+def ego_graph(G, n, radius=1, center=True, undirected=False, is_weighted=False):
+ # TODO: should we have an option to keep the output matrix the same size?
+ if undirected and G.is_directed():
+ # NOT COVERED
+ G2 = G.to_undirected()
+ else:
+ G2 = G
+ if is_weighted:
+ v = single_source_bellman_ford_path_length(G2, n, cutoff=radius)
+ else:
+ v = _plain_bfs(G2, n, cutoff=radius)
+ if not center:
+ del v[G._key_to_id[n]]
+
+ indices, _ = v.to_coo(values=False)
+ A = G._A[indices, indices].new(name="ego")
+ key_to_id = G.renumber_key_to_id(indices.tolist())
+ return type(G)(A, key_to_id=key_to_id)
diff --git a/graphblas_algorithms/interface.py b/graphblas_algorithms/interface.py
index 206d19c..94f02a6 100644
--- a/graphblas_algorithms/interface.py
+++ b/graphblas_algorithms/interface.py
@@ -45,6 +45,8 @@ class Dispatcher:
descendants = nxapi.dag.descendants
# Dominating
is_dominating_set = nxapi.dominating.is_dominating_set
+ # Generators
+ ego_graph = nxapi.generators.ego.ego_graph
# Isolate
is_isolate = nxapi.isolate.is_isolate
isolates = nxapi.isolate.isolates
diff --git a/graphblas_algorithms/nxapi/__init__.py b/graphblas_algorithms/nxapi/__init__.py
index 5ddc1fa..2d36017 100644
--- a/graphblas_algorithms/nxapi/__init__.py
+++ b/graphblas_algorithms/nxapi/__init__.py
@@ -7,6 +7,7 @@
from .cuts import *
from .dag import *
from .dominating import *
+from .generators import *
from .isolate import *
from .link_analysis import *
from .operators import *
@@ -23,6 +24,7 @@
from . import cluster
from . import community
from . import components
+from . import generators
from . import link_analysis
from . import operators
from . import shortest_paths
diff --git a/graphblas_algorithms/nxapi/generators/__init__.py b/graphblas_algorithms/nxapi/generators/__init__.py
new file mode 100644
index 0000000..65a6526
--- /dev/null
+++ b/graphblas_algorithms/nxapi/generators/__init__.py
@@ -0,0 +1 @@
+from .ego import *
diff --git a/graphblas_algorithms/nxapi/generators/ego.py b/graphblas_algorithms/nxapi/generators/ego.py
new file mode 100644
index 0000000..e591cb3
--- /dev/null
+++ b/graphblas_algorithms/nxapi/generators/ego.py
@@ -0,0 +1,11 @@
+from graphblas_algorithms import generators
+from graphblas_algorithms.classes.digraph import to_graph
+
+__all__ = ["ego_graph"]
+
+
+def ego_graph(G, n, radius=1, center=True, undirected=False, distance=None):
+ G = to_graph(G, weight=distance)
+ return generators.ego_graph(
+ G, n, radius=radius, center=center, undirected=undirected, is_weighted=distance is not None
+ )
diff --git a/graphblas_algorithms/tests/test_match_nx.py b/graphblas_algorithms/tests/test_match_nx.py
index c50896f..490d1d7 100644
--- a/graphblas_algorithms/tests/test_match_nx.py
+++ b/graphblas_algorithms/tests/test_match_nx.py
@@ -159,3 +159,35 @@ def test_dispatched_funcs_in_nxapi(nx_names_to_info, gb_names_to_info):
print(" ", ":".join(path.rsplit(".", 1)))
if failing: # pragma: no cover
raise AssertionError
+
+
+def test_print_dispatched_not_implemented(nx_names_to_info, gb_names_to_info):
+ """It may be informative to see the results from this to identify functions to implement.
+
+ $ pytest -s -k test_print_dispatched_not_implemented
+ """
+ not_implemented = nx_names_to_info.keys() - gb_names_to_info.keys()
+ fullnames = {next(iter(nx_names_to_info[name])).fullname for name in not_implemented}
+ print()
+ print("=================================================================================")
+ print("Functions dispatched in NetworkX that ARE NOT implemented in graphblas-algorithms")
+ print("---------------------------------------------------------------------------------")
+ for i, name in enumerate(sorted(fullnames)):
+ print(i, name)
+ print("=================================================================================")
+
+
+def test_print_dispatched_implemented(nx_names_to_info, gb_names_to_info):
+ """It may be informative to see the results from this to identify implemented functions.
+
+ $ pytest -s -k test_print_dispatched_implemented
+ """
+ implemented = nx_names_to_info.keys() & gb_names_to_info.keys()
+ fullnames = {next(iter(nx_names_to_info[name])).fullname for name in implemented}
+ print()
+ print("=============================================================================")
+ print("Functions dispatched in NetworkX that ARE implemented in graphblas-algorithms")
+ print("-----------------------------------------------------------------------------")
+ for i, name in enumerate(sorted(fullnames)):
+ print(i, name)
+ print("=============================================================================")
diff --git a/pyproject.toml b/pyproject.toml
index df17600..1772aa2 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -36,7 +36,7 @@ keywords = [
"math",
]
classifiers = [
- "Development Status :: 3 - Alpha",
+ "Development Status :: 4 - Beta",
"License :: OSI Approved :: Apache Software License",
"Operating System :: MacOS :: MacOS X",
"Operating System :: POSIX :: Linux",
@@ -46,6 +46,7 @@ classifiers = [
"Programming Language :: Python :: 3.8",
"Programming Language :: Python :: 3.9",
"Programming Language :: Python :: 3.10",
+ "Programming Language :: Python :: 3.11",
"Programming Language :: Python :: 3 :: Only",
"Intended Audience :: Developers",
"Intended Audience :: Other Audience",
@@ -78,12 +79,8 @@ test = [
"setuptools",
"tomli",
]
-complete = [
- "pytest",
- "networkx >=3.0",
- "scipy >=1.8",
- "setuptools",
- "tomli",
+all = [
+ "graphblas-algorithms[test]",
]
[tool.setuptools]
@@ -104,10 +101,12 @@ packages = [
"graphblas_algorithms.algorithms.tests",
"graphblas_algorithms.algorithms.traversal",
"graphblas_algorithms.classes",
+ "graphblas_algorithms.generators",
"graphblas_algorithms.nxapi",
"graphblas_algorithms.nxapi.centrality",
"graphblas_algorithms.nxapi.community",
"graphblas_algorithms.nxapi.components",
+ "graphblas_algorithms.nxapi.generators",
"graphblas_algorithms.nxapi.link_analysis",
"graphblas_algorithms.nxapi.operators",
"graphblas_algorithms.nxapi.shortest_paths",
### Installation
```
@@ -151,6 +151,8 @@ dispatch pattern shown above.
- descendants
- Dominating
- is_dominating_set
+- Generators
+ - ego_graph
- Isolate
- is_isolate
- isolates
diff --git a/graphblas_algorithms/__init__.py b/graphblas_algorithms/__init__.py
index f9bbcf4..09418f5 100644
--- a/graphblas_algorithms/__init__.py
+++ b/graphblas_algorithms/__init__.py
@@ -3,6 +3,7 @@
from .classes import *
from .algorithms import * # isort:skip
+from .generators import * # isort:skip
try:
__version__ = importlib.metadata.version("graphblas-algorithms")
diff --git a/graphblas_algorithms/algorithms/_bfs.py b/graphblas_algorithms/algorithms/_bfs.py
new file mode 100644
index 0000000..31674ce
--- /dev/null
+++ b/graphblas_algorithms/algorithms/_bfs.py
@@ -0,0 +1,150 @@
+"""BFS routines used by other algorithms"""
+
+import numpy as np
+from graphblas import Matrix, Vector, binary, replace, unary
+from graphblas.semiring import any_pair
+
+
+def _get_cutoff(n, cutoff):
+ if cutoff is None or cutoff >= n:
+ return n # Everything
+ return cutoff + 1 # Inclusive
+
+
+def _plain_bfs(G, source, *, cutoff=None):
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q = Vector(bool, n, name="q")
+ v[index] = True
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for _i in range(1, cutoff):
+ q(~v.S, replace) << any_pair_bool(q @ A)
+ if q.nvals == 0:
+ break
+ v(q.S) << True
+ return v
+
+
+def _bfs_level(G, source, cutoff=None, *, transpose=False, dtype=int):
+ if dtype == bool:
+ dtype = int
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ if transpose and G.is_directed():
+ A = A.T # TODO: should we use "AT" instead?
+ n = A.nrows
+ v = Vector(dtype, n, name="bfs_level")
+ q = Vector(bool, n, name="q")
+ v[index] = 0
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for i in range(1, cutoff):
+ q(~v.S, replace) << any_pair_bool(q @ A)
+ if q.nvals == 0:
+ break
+ v(q.S) << i
+ return v
+
+
+def _bfs_levels(G, nodes, cutoff=None, *, dtype=int):
+ if dtype == bool:
+ dtype = int
+ A = G.get_property("offdiag")
+ n = A.nrows
+ if nodes is None:
+ # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
+ D = Vector(dtype, n, name="bfs_levels_vector")
+ D << 0
+ D = D.diag(name="bfs_levels")
+ else:
+ ids = G.list_to_ids(nodes)
+ D = Matrix.from_coo(
+ np.arange(len(ids), dtype=np.uint64),
+ ids,
+ 0,
+ dtype,
+ nrows=len(ids),
+ ncols=n,
+ name="bfs_levels",
+ )
+ Q = unary.one[bool](D).new(name="Q")
+ any_pair_bool = any_pair[bool]
+ cutoff = _get_cutoff(n, cutoff)
+ for i in range(1, cutoff):
+ Q(~D.S, replace) << any_pair_bool(Q @ A)
+ if Q.nvals == 0:
+ break
+ D(Q.S) << i
+ return D
+
+
+# TODO: benchmark this and the version commented out below
+def _plain_bfs_bidirectional(G, source):
+ # Bi-directional BFS w/o symmetrizing the adjacency matrix
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ # XXX: should we use `AT` if available?
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q_out = Vector(bool, n, name="q_out")
+ q_in = Vector(bool, n, name="q_in")
+ v[index] = True
+ q_in[index] = True
+ any_pair_bool = any_pair[bool]
+ is_out_empty = True
+ is_in_empty = False
+ for _i in range(1, n):
+ # Traverse out-edges from the most recent `q_in` and `q_out`
+ if is_out_empty:
+ q_out(~v.S) << any_pair_bool(q_in @ A)
+ else:
+ q_out << binary.any(q_out | q_in)
+ q_out(~v.S, replace) << any_pair_bool(q_out @ A)
+ is_out_empty = q_out.nvals == 0
+ if not is_out_empty:
+ v(q_out.S) << True
+ elif is_in_empty:
+ break
+ # Traverse in-edges from the most recent `q_in` and `q_out`
+ if is_in_empty:
+ q_in(~v.S) << any_pair_bool(A @ q_out)
+ else:
+ q_in << binary.any(q_out | q_in)
+ q_in(~v.S, replace) << any_pair_bool(A @ q_in)
+ is_in_empty = q_in.nvals == 0
+ if not is_in_empty:
+ v(q_in.S) << True
+ elif is_out_empty:
+ break
+ return v
+
+
+"""
+def _plain_bfs_bidirectional(G, source):
+ # Bi-directional BFS w/o symmetrizing the adjacency matrix
+ index = G._key_to_id[source]
+ A = G.get_property("offdiag")
+ n = A.nrows
+ v = Vector(bool, n, name="bfs_plain")
+ q = Vector(bool, n, name="q")
+ q2 = Vector(bool, n, name="q_2")
+ v[index] = True
+ q[index] = True
+ any_pair_bool = any_pair[bool]
+ for _i in range(1, n):
+ q2(~v.S, replace) << any_pair_bool(q @ A)
+ v(q2.S) << True
+ q(~v.S, replace) << any_pair_bool(A @ q)
+ if q.nvals == 0:
+ if q2.nvals == 0:
+ break
+ q, q2 = q2, q
+ elif q2.nvals != 0:
+ q << binary.any(q | q2)
+ return v
+"""
diff --git a/graphblas_algorithms/algorithms/centrality/eigenvector.py b/graphblas_algorithms/algorithms/centrality/eigenvector.py
index 5172f61..e9385f3 100644
--- a/graphblas_algorithms/algorithms/centrality/eigenvector.py
+++ b/graphblas_algorithms/algorithms/centrality/eigenvector.py
@@ -1,11 +1,7 @@
from graphblas import Vector
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import (
- ConvergenceFailure,
- GraphBlasAlgorithmException,
- PointlessConcept,
-)
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure, GraphBlasAlgorithmException, PointlessConcept
__all__ = ["eigenvector_centrality"]
diff --git a/graphblas_algorithms/algorithms/centrality/katz.py b/graphblas_algorithms/algorithms/centrality/katz.py
index 78de982..8087e85 100644
--- a/graphblas_algorithms/algorithms/centrality/katz.py
+++ b/graphblas_algorithms/algorithms/centrality/katz.py
@@ -2,11 +2,8 @@
from graphblas.core.utils import output_type
from graphblas.semiring import plus_first, plus_times
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import (
- ConvergenceFailure,
- GraphBlasAlgorithmException,
-)
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure, GraphBlasAlgorithmException
__all__ = ["katz_centrality"]
diff --git a/graphblas_algorithms/algorithms/components/connected.py b/graphblas_algorithms/algorithms/components/connected.py
index 37c0fc9..fb2f678 100644
--- a/graphblas_algorithms/algorithms/components/connected.py
+++ b/graphblas_algorithms/algorithms/components/connected.py
@@ -1,7 +1,5 @@
-from graphblas import Vector, replace
-from graphblas.semiring import any_pair
-
-from graphblas_algorithms.algorithms.exceptions import PointlessConcept
+from .._bfs import _plain_bfs
+from ..exceptions import PointlessConcept
def is_connected(G):
@@ -12,20 +10,3 @@ def is_connected(G):
def node_connected_component(G, n):
return _plain_bfs(G, n)
-
-
-def _plain_bfs(G, source):
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q = Vector(bool, n, name="q")
- v[index] = True
- q[index] = True
- any_pair_bool = any_pair[bool]
- for _i in range(1, n):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << True
- return v
diff --git a/graphblas_algorithms/algorithms/components/weakly_connected.py b/graphblas_algorithms/algorithms/components/weakly_connected.py
index eb3dc75..99eba78 100644
--- a/graphblas_algorithms/algorithms/components/weakly_connected.py
+++ b/graphblas_algorithms/algorithms/components/weakly_connected.py
@@ -1,77 +1,8 @@
-from graphblas import Vector, binary, replace
-from graphblas.semiring import any_pair
-
-from graphblas_algorithms.algorithms.exceptions import PointlessConcept
+from .._bfs import _plain_bfs_bidirectional
+from ..exceptions import PointlessConcept
def is_weakly_connected(G):
if len(G) == 0:
raise PointlessConcept("Connectivity is undefined for the null graph.")
- return _plain_bfs(G, next(iter(G))).nvals == len(G)
-
-
-# TODO: benchmark this and the version commented out below
-def _plain_bfs(G, source):
- # Bi-directional BFS w/o symmetrizing the adjacency matrix
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- # XXX: should we use `AT` if available?
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q_out = Vector(bool, n, name="q_out")
- q_in = Vector(bool, n, name="q_in")
- v[index] = True
- q_in[index] = True
- any_pair_bool = any_pair[bool]
- is_out_empty = True
- is_in_empty = False
- for _i in range(1, n):
- # Traverse out-edges from the most recent `q_in` and `q_out`
- if is_out_empty:
- q_out(~v.S) << any_pair_bool(q_in @ A)
- else:
- q_out << binary.any(q_out | q_in)
- q_out(~v.S, replace) << any_pair_bool(q_out @ A)
- is_out_empty = q_out.nvals == 0
- if not is_out_empty:
- v(q_out.S) << True
- elif is_in_empty:
- break
- # Traverse in-edges from the most recent `q_in` and `q_out`
- if is_in_empty:
- q_in(~v.S) << any_pair_bool(A @ q_out)
- else:
- q_in << binary.any(q_out | q_in)
- q_in(~v.S, replace) << any_pair_bool(A @ q_in)
- is_in_empty = q_in.nvals == 0
- if not is_in_empty:
- v(q_in.S) << True
- elif is_out_empty:
- break
- return v
-
-
-"""
-def _plain_bfs(G, source):
- # Bi-directional BFS w/o symmetrizing the adjacency matrix
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(bool, n, name="bfs_plain")
- q = Vector(bool, n, name="q")
- q2 = Vector(bool, n, name="q_2")
- v[index] = True
- q[index] = True
- any_pair_bool = any_pair[bool]
- for _i in range(1, n):
- q2(~v.S, replace) << any_pair_bool(q @ A)
- v(q2.S) << True
- q(~v.S, replace) << any_pair_bool(A @ q)
- if q.nvals == 0:
- if q2.nvals == 0:
- break
- q, q2 = q2, q
- elif q2.nvals != 0:
- q << binary.any(q | q2)
- return v
-"""
+ return _plain_bfs_bidirectional(G, next(iter(G))).nvals == len(G)
diff --git a/graphblas_algorithms/algorithms/core.py b/graphblas_algorithms/algorithms/core.py
index 8133c71..a6ff26d 100644
--- a/graphblas_algorithms/algorithms/core.py
+++ b/graphblas_algorithms/algorithms/core.py
@@ -1,11 +1,12 @@
from graphblas import Matrix, monoid, replace, select, semiring
-from graphblas_algorithms.classes.graph import Graph
+from graphblas_algorithms import Graph
__all__ = ["k_truss"]
def k_truss(G: Graph, k) -> Graph:
+ # TODO: should we have an option to keep the output matrix the same size?
# Ignore self-edges
S = G.get_property("offdiag")
@@ -32,6 +33,5 @@ def k_truss(G: Graph, k) -> Graph:
Ktruss = C[indices, indices].new()
# Convert back to networkx graph with correct node ids
- keys = G.list_to_keys(indices)
- key_to_id = dict(zip(keys, range(len(indices))))
+ key_to_id = G.renumber_key_to_id(indices.tolist())
return Graph(Ktruss, key_to_id=key_to_id)
diff --git a/graphblas_algorithms/algorithms/link_analysis/hits_alg.py b/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
index 515806e..662ac14 100644
--- a/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
+++ b/graphblas_algorithms/algorithms/link_analysis/hits_alg.py
@@ -1,7 +1,7 @@
from graphblas import Vector
-from graphblas_algorithms.algorithms._helpers import is_converged, normalize
-from graphblas_algorithms.algorithms.exceptions import ConvergenceFailure
+from .._helpers import is_converged, normalize
+from ..exceptions import ConvergenceFailure
__all__ = ["hits"]
diff --git a/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py b/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
index 518c09f..d665e98 100644
--- a/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
+++ b/graphblas_algorithms/algorithms/link_analysis/pagerank_alg.py
@@ -3,8 +3,9 @@
from graphblas.semiring import plus_first, plus_times
from graphblas_algorithms import Graph
-from graphblas_algorithms.algorithms._helpers import is_converged
-from graphblas_algorithms.algorithms.exceptions import ConvergenceFailure
+
+from .._helpers import is_converged
+from ..exceptions import ConvergenceFailure
__all__ = ["pagerank", "google_matrix"]
diff --git a/graphblas_algorithms/algorithms/shortest_paths/unweighted.py b/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
index 3c8243f..5062e87 100644
--- a/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
+++ b/graphblas_algorithms/algorithms/shortest_paths/unweighted.py
@@ -1,6 +1,6 @@
-import numpy as np
-from graphblas import Matrix, Vector, replace, unary
-from graphblas.semiring import any_pair
+from graphblas import Matrix
+
+from .._bfs import _bfs_level, _bfs_levels
__all__ = [
"single_source_shortest_path_length",
@@ -25,59 +25,3 @@ def all_pairs_shortest_path_length(G, cutoff=None, *, nodes=None, expand_output=
rv[ids, :] = D
return rv
return D
-
-
-def _bfs_level(G, source, cutoff, *, transpose=False):
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- if transpose and G.is_directed():
- A = A.T # TODO: should we use "AT" instead?
- n = A.nrows
- v = Vector(int, n, name="bfs_unweighted")
- q = Vector(bool, n, name="q")
- v[index] = 0
- q[index] = True
- any_pair_bool = any_pair[bool]
- if cutoff is None or cutoff >= n:
- cutoff = n # Everything
- else:
- cutoff += 1 # Inclusive
- for i in range(1, cutoff):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << i
- return v
-
-
-def _bfs_levels(G, nodes, cutoff):
- A = G.get_property("offdiag")
- n = A.nrows
- if nodes is None:
- # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
- D = Vector(int, n, name="bfs_unweighted_vector")
- D << 0
- D = D.diag(name="bfs_unweighted")
- else:
- ids = G.list_to_ids(nodes)
- D = Matrix.from_coo(
- np.arange(len(ids), dtype=np.uint64),
- ids,
- 0,
- int,
- nrows=len(ids),
- ncols=n,
- name="bfs_unweighted",
- )
- Q = unary.one[bool](D).new(name="Q")
- any_pair_bool = any_pair[bool]
- if cutoff is None or cutoff >= n:
- cutoff = n # Everything
- else:
- cutoff += 1 # Inclusive
- for i in range(1, cutoff):
- Q(~D.S, replace) << any_pair_bool(Q @ A)
- if Q.nvals == 0:
- break
- D(Q.S) << i
- return D
diff --git a/graphblas_algorithms/algorithms/shortest_paths/weighted.py b/graphblas_algorithms/algorithms/shortest_paths/weighted.py
index 8e6efef..fddf672 100644
--- a/graphblas_algorithms/algorithms/shortest_paths/weighted.py
+++ b/graphblas_algorithms/algorithms/shortest_paths/weighted.py
@@ -2,6 +2,7 @@
from graphblas import Matrix, Vector, binary, monoid, replace, select, unary
from graphblas.semiring import any_pair, min_plus
+from .._bfs import _bfs_level, _bfs_levels
from ..exceptions import Unbounded
__all__ = [
@@ -11,14 +12,16 @@
]
-def single_source_bellman_ford_path_length(G, source):
+def single_source_bellman_ford_path_length(G, source, *, cutoff=None):
# No need for `is_weighted=` keyword, b/c this is assumed to be weighted (I think)
index = G._key_to_id[source]
if G.get_property("is_iso"):
# If the edges are iso-valued (and positive), then we can simply do level BFS
is_negative, iso_value = G.get_properties("has_negative_edges+ iso_value")
if not is_negative:
- d = _bfs_level(G, source, dtype=iso_value.dtype)
+ if cutoff is not None:
+ cutoff = int(cutoff // iso_value)
+ d = _bfs_level(G, source, cutoff, dtype=iso_value.dtype)
if iso_value != 1:
d *= iso_value
return d
@@ -49,6 +52,8 @@ def single_source_bellman_ford_path_length(G, source):
# `cur` is the current frontier of values that improved in the previous iteration.
# This means that in this iteration we drop values from `cur` that are not better.
cur << min_plus(cur @ A)
+ if cutoff is not None:
+ cur << select.valuele(cur, cutoff)
# Mask is True where cur not in d or cur < d
mask << one(cur)
@@ -63,6 +68,8 @@ def single_source_bellman_ford_path_length(G, source):
else:
# Check for negative cycle when for loop completes without breaking
cur << min_plus(cur @ A)
+ if cutoff is not None:
+ cur << select.valuele(cur, cutoff)
mask << binary.lt(cur & d)
if mask.reduce(monoid.lor):
raise Unbounded("Negative cycle detected.")
@@ -157,56 +164,6 @@ def bellman_ford_path_lengths(G, nodes=None, *, expand_output=False):
return D
-def _bfs_level(G, source, *, dtype=int):
- if dtype == bool:
- dtype = int
- index = G._key_to_id[source]
- A = G.get_property("offdiag")
- n = A.nrows
- v = Vector(dtype, n, name="bfs_level")
- q = Vector(bool, n, name="q")
- v[index] = 0
- q[index] = True
- any_pair_bool = any_pair[bool]
- for i in range(1, n):
- q(~v.S, replace) << any_pair_bool(q @ A)
- if q.nvals == 0:
- break
- v(q.S) << i
- return v
-
-
-def _bfs_levels(G, nodes=None, *, dtype=int):
- if dtype == bool:
- dtype = int
- A = G.get_property("offdiag")
- n = A.nrows
- if nodes is None:
- # TODO: `D = Vector.from_scalar(0, n, dtype).diag()`
- D = Vector(dtype, n, name="bfs_levels_vector")
- D << 0
- D = D.diag(name="bfs_levels")
- else:
- ids = G.list_to_ids(nodes)
- D = Matrix.from_coo(
- np.arange(len(ids), dtype=np.uint64),
- ids,
- 0,
- dtype,
- nrows=len(ids),
- ncols=n,
- name="bfs_levels",
- )
- Q = unary.one[bool](D).new(name="Q")
- any_pair_bool = any_pair[bool]
- for i in range(1, n):
- Q(~D.S, replace) << any_pair_bool(Q @ A)
- if Q.nvals == 0:
- break
- D(Q.S) << i
- return D
-
-
def negative_edge_cycle(G):
# TODO: use a heuristic to try to stop early
if G.is_directed():
diff --git a/graphblas_algorithms/algorithms/triads.py b/graphblas_algorithms/algorithms/triads.py
index 54702c7..e6ec2be 100644
--- a/graphblas_algorithms/algorithms/triads.py
+++ b/graphblas_algorithms/algorithms/triads.py
@@ -1,5 +1,4 @@
-from graphblas_algorithms.classes.digraph import DiGraph
-from graphblas_algorithms.classes.graph import Graph
+from graphblas_algorithms import DiGraph, Graph
__all__ = ["is_triad"]
diff --git a/graphblas_algorithms/classes/_utils.py b/graphblas_algorithms/classes/_utils.py
index 92febc5..65ae010 100644
--- a/graphblas_algorithms/classes/_utils.py
+++ b/graphblas_algorithms/classes/_utils.py
@@ -250,3 +250,12 @@ def _cacheit(self, key, func, *args, **kwargs):
if key not in self._cache:
self._cache[key] = func(*args, **kwargs)
return self._cache[key]
+
+
+def renumber_key_to_id(self, indices):
+ """Create `key_to_id` for e.g. a subgraph with node ids from `indices`"""
+ id_to_key = self.id_to_key
+ return {id_to_key[index]: i for i, index in enumerate(indices)}
+ # Alternative (about the same performance)
+ # keys = self.list_to_keys(indices)
+ # return dict(zip(keys, range(len(indices))))
diff --git a/graphblas_algorithms/classes/digraph.py b/graphblas_algorithms/classes/digraph.py
index 83e7356..bae66ae 100644
--- a/graphblas_algorithms/classes/digraph.py
+++ b/graphblas_algorithms/classes/digraph.py
@@ -553,6 +553,7 @@ def __init__(self, incoming_graph_data=None, *, key_to_id=None, **attr):
vector_to_nodeset = _utils.vector_to_nodeset
vector_to_set = _utils.vector_to_set
_cacheit = _utils._cacheit
+ renumber_key_to_id = _utils.renumber_key_to_id
# NetworkX methods
def to_directed_class(self):
@@ -598,6 +599,16 @@ def is_multigraph(self):
def is_directed(self):
return True
+ def to_undirected(self, reciprocal=False, as_view=False, *, name=None):
+ if as_view:
+ raise NotImplementedError("`as_vew=True` is not implemented in `G.to_undirected`")
+ A = self._A
+ if reciprocal:
+ B = binary.any(A & A.T).new(name=name)
+ else:
+ B = binary.any(A | A.T).new(name=name)
+ return Graph(B, key_to_id=self._key_to_id)
+
class MultiDiGraph(DiGraph):
def is_multigraph(self):
diff --git a/graphblas_algorithms/classes/graph.py b/graphblas_algorithms/classes/graph.py
index 03a2893..06f82be 100644
--- a/graphblas_algorithms/classes/graph.py
+++ b/graphblas_algorithms/classes/graph.py
@@ -401,6 +401,7 @@ def __init__(self, incoming_graph_data=None, *, key_to_id=None, **attr):
vector_to_nodeset = _utils.vector_to_nodeset
vector_to_set = _utils.vector_to_set
_cacheit = _utils._cacheit
+ renumber_key_to_id = _utils.renumber_key_to_id
# NetworkX methods
def to_directed_class(self):
diff --git a/graphblas_algorithms/generators/__init__.py b/graphblas_algorithms/generators/__init__.py
new file mode 100644
index 0000000..65a6526
--- /dev/null
+++ b/graphblas_algorithms/generators/__init__.py
@@ -0,0 +1 @@
+from .ego import *
diff --git a/graphblas_algorithms/generators/ego.py b/graphblas_algorithms/generators/ego.py
new file mode 100644
index 0000000..26e9cf9
--- /dev/null
+++ b/graphblas_algorithms/generators/ego.py
@@ -0,0 +1,24 @@
+from ..algorithms.components.connected import _plain_bfs
+from ..algorithms.shortest_paths.weighted import single_source_bellman_ford_path_length
+
+__all__ = ["ego_graph"]
+
+
+def ego_graph(G, n, radius=1, center=True, undirected=False, is_weighted=False):
+ # TODO: should we have an option to keep the output matrix the same size?
+ if undirected and G.is_directed():
+ # NOT COVERED
+ G2 = G.to_undirected()
+ else:
+ G2 = G
+ if is_weighted:
+ v = single_source_bellman_ford_path_length(G2, n, cutoff=radius)
+ else:
+ v = _plain_bfs(G2, n, cutoff=radius)
+ if not center:
+ del v[G._key_to_id[n]]
+
+ indices, _ = v.to_coo(values=False)
+ A = G._A[indices, indices].new(name="ego")
+ key_to_id = G.renumber_key_to_id(indices.tolist())
+ return type(G)(A, key_to_id=key_to_id)
diff --git a/graphblas_algorithms/interface.py b/graphblas_algorithms/interface.py
index 206d19c..94f02a6 100644
--- a/graphblas_algorithms/interface.py
+++ b/graphblas_algorithms/interface.py
@@ -45,6 +45,8 @@ class Dispatcher:
descendants = nxapi.dag.descendants
# Dominating
is_dominating_set = nxapi.dominating.is_dominating_set
+ # Generators
+ ego_graph = nxapi.generators.ego.ego_graph
# Isolate
is_isolate = nxapi.isolate.is_isolate
isolates = nxapi.isolate.isolates
diff --git a/graphblas_algorithms/nxapi/__init__.py b/graphblas_algorithms/nxapi/__init__.py
index 5ddc1fa..2d36017 100644
--- a/graphblas_algorithms/nxapi/__init__.py
+++ b/graphblas_algorithms/nxapi/__init__.py
@@ -7,6 +7,7 @@
from .cuts import *
from .dag import *
from .dominating import *
+from .generators import *
from .isolate import *
from .link_analysis import *
from .operators import *
@@ -23,6 +24,7 @@
from . import cluster
from . import community
from . import components
+from . import generators
from . import link_analysis
from . import operators
from . import shortest_paths
diff --git a/graphblas_algorithms/nxapi/generators/__init__.py b/graphblas_algorithms/nxapi/generators/__init__.py
new file mode 100644
index 0000000..65a6526
--- /dev/null
+++ b/graphblas_algorithms/nxapi/generators/__init__.py
@@ -0,0 +1 @@
+from .ego import *
diff --git a/graphblas_algorithms/nxapi/generators/ego.py b/graphblas_algorithms/nxapi/generators/ego.py
new file mode 100644
index 0000000..e591cb3
--- /dev/null
+++ b/graphblas_algorithms/nxapi/generators/ego.py
@@ -0,0 +1,11 @@
+from graphblas_algorithms import generators
+from graphblas_algorithms.classes.digraph import to_graph
+
+__all__ = ["ego_graph"]
+
+
+def ego_graph(G, n, radius=1, center=True, undirected=False, distance=None):
+ G = to_graph(G, weight=distance)
+ return generators.ego_graph(
+ G, n, radius=radius, center=center, undirected=undirected, is_weighted=distance is not None
+ )
diff --git a/graphblas_algorithms/tests/test_match_nx.py b/graphblas_algorithms/tests/test_match_nx.py
index c50896f..490d1d7 100644
--- a/graphblas_algorithms/tests/test_match_nx.py
+++ b/graphblas_algorithms/tests/test_match_nx.py
@@ -159,3 +159,35 @@ def test_dispatched_funcs_in_nxapi(nx_names_to_info, gb_names_to_info):
print(" ", ":".join(path.rsplit(".", 1)))
if failing: # pragma: no cover
raise AssertionError
+
+
+def test_print_dispatched_not_implemented(nx_names_to_info, gb_names_to_info):
+ """It may be informative to see the results from this to identify functions to implement.
+
+ $ pytest -s -k test_print_dispatched_not_implemented
+ """
+ not_implemented = nx_names_to_info.keys() - gb_names_to_info.keys()
+ fullnames = {next(iter(nx_names_to_info[name])).fullname for name in not_implemented}
+ print()
+ print("=================================================================================")
+ print("Functions dispatched in NetworkX that ARE NOT implemented in graphblas-algorithms")
+ print("---------------------------------------------------------------------------------")
+ for i, name in enumerate(sorted(fullnames)):
+ print(i, name)
+ print("=================================================================================")
+
+
+def test_print_dispatched_implemented(nx_names_to_info, gb_names_to_info):
+ """It may be informative to see the results from this to identify implemented functions.
+
+ $ pytest -s -k test_print_dispatched_implemented
+ """
+ implemented = nx_names_to_info.keys() & gb_names_to_info.keys()
+ fullnames = {next(iter(nx_names_to_info[name])).fullname for name in implemented}
+ print()
+ print("=============================================================================")
+ print("Functions dispatched in NetworkX that ARE implemented in graphblas-algorithms")
+ print("-----------------------------------------------------------------------------")
+ for i, name in enumerate(sorted(fullnames)):
+ print(i, name)
+ print("=============================================================================")
diff --git a/pyproject.toml b/pyproject.toml
index df17600..1772aa2 100644
--- a/pyproject.toml
+++ b/pyproject.toml
@@ -36,7 +36,7 @@ keywords = [
"math",
]
classifiers = [
- "Development Status :: 3 - Alpha",
+ "Development Status :: 4 - Beta",
"License :: OSI Approved :: Apache Software License",
"Operating System :: MacOS :: MacOS X",
"Operating System :: POSIX :: Linux",
@@ -46,6 +46,7 @@ classifiers = [
"Programming Language :: Python :: 3.8",
"Programming Language :: Python :: 3.9",
"Programming Language :: Python :: 3.10",
+ "Programming Language :: Python :: 3.11",
"Programming Language :: Python :: 3 :: Only",
"Intended Audience :: Developers",
"Intended Audience :: Other Audience",
@@ -78,12 +79,8 @@ test = [
"setuptools",
"tomli",
]
-complete = [
- "pytest",
- "networkx >=3.0",
- "scipy >=1.8",
- "setuptools",
- "tomli",
+all = [
+ "graphblas-algorithms[test]",
]
[tool.setuptools]
@@ -104,10 +101,12 @@ packages = [
"graphblas_algorithms.algorithms.tests",
"graphblas_algorithms.algorithms.traversal",
"graphblas_algorithms.classes",
+ "graphblas_algorithms.generators",
"graphblas_algorithms.nxapi",
"graphblas_algorithms.nxapi.centrality",
"graphblas_algorithms.nxapi.community",
"graphblas_algorithms.nxapi.components",
+ "graphblas_algorithms.nxapi.generators",
"graphblas_algorithms.nxapi.link_analysis",
"graphblas_algorithms.nxapi.operators",
"graphblas_algorithms.nxapi.shortest_paths",