A script can calculate residues-residues contact scores (rrcs) of a trajectory file created by GROMACs.
This project is just a script, so it doesn't require overly complicated installation steps. You only need to install the relevant dependencies using pip or pipenv in your working environment to run it:
pip install -r requirements.txt
One of the following: After Put the script in dictionary example
python3 gmx_rrcs.py example.txt
Among them, the file "example.txt" is the input parameter format. An example is shown below. The relevant parameters have a default value, while the topology file (top) and trajectory (traj) file are required.
r_min = 3.23 ;Specify the minimum distance for atomic contacts. The default value is 3.23 Å.
r_max = 4.63 ;Specify the maximum distance for atomic contacts. The default value is 4.63 Å.
bt = 0.0 ;The starting time for the calculation, in units of ps. The default value is 0.0 ps.
et = 100.0 ;The end time for the calculation, in units of ps. The default value is all.
dt = 1.0 ;The frequency for calculating each frame in units of ps, by default it is set to calculate every frame.
res = index.txt;The file name of the residue index file.
top = md0.pdb ;The file name of the topology file. It must be the PDB file of the first frame in the trajectory file.
traj = md0.xtc ;The file name of the trajectory file.
Because we often don't need to calculate rrcs for every residue pair, which would greatly reduce the running efficiency. You can input an "index.txt" file that contains the residues (pairs) that need to be observed, and enter the file name in the parameter file. If the residue index file is not specified, all residues will be observed by default.
An example residue index file is here:
1 $ 2-5 13 16 ; This indicates the calculation of rrcs between residue 1 and residues 2-5, 13, and 16. The residue number before the "$" can only one.
23-27 ; If there is no "$", then it means calculating the rrcs between each residue pair indicated by the data in this row.
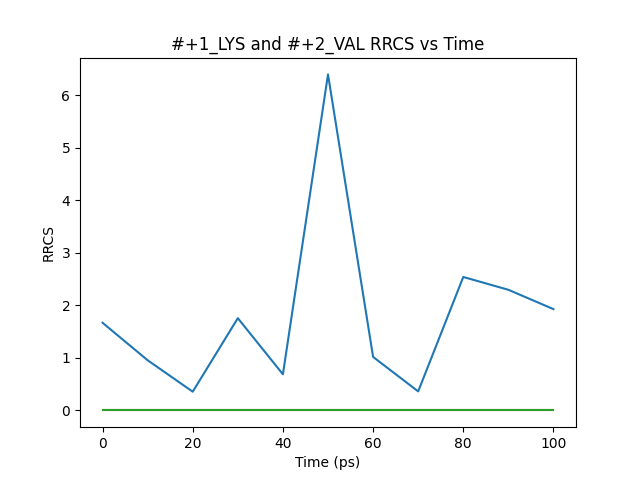
We have adopted a simple output file format. It creates a folder with the same name as the parameter file, and outputs the rrcs and time variation graphs (in PNG format) for each residue pair in the folder.
This may not be of publication quality, but it provides the necessary variation information. In addition, the script also outputs a file in XVG format, which allows users to process the data using other plotting software (such as gnuplot).
If the files you output are as follows, then your run is considered successful: