-
Notifications
You must be signed in to change notification settings - Fork 15
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
xarray writing mfdataset results in incorrect data when not using manual encoding #239
Comments
|
This is solved with dfmt.prevent_dtype_int() which is used in dfmt.merge_meteofiles(): The new examplefile is preprocess_merge_meteofiles.py |
|
Solved with above, but another workaround is removing the |
|
Alternatively, re-compute scaling/offset like suggested in ArcticSnow/TopoPyScale#60 (comment) Implementation: https://github.com/ArcticSnow/TopoPyScale/blob/494f4e7ea17830ba3d23627bf22ee200a6c4f082/TopoPyScale/topo_export.py#L21 |
|
Recompute issue created: #269 |
|
Zipping might be easier and more generic, however, some encoding has to be altered when doing that (for each variable). |
|
GTSM fou ncfiles were easily compressed and without performance reduction ( For era5 it would be something like this: |


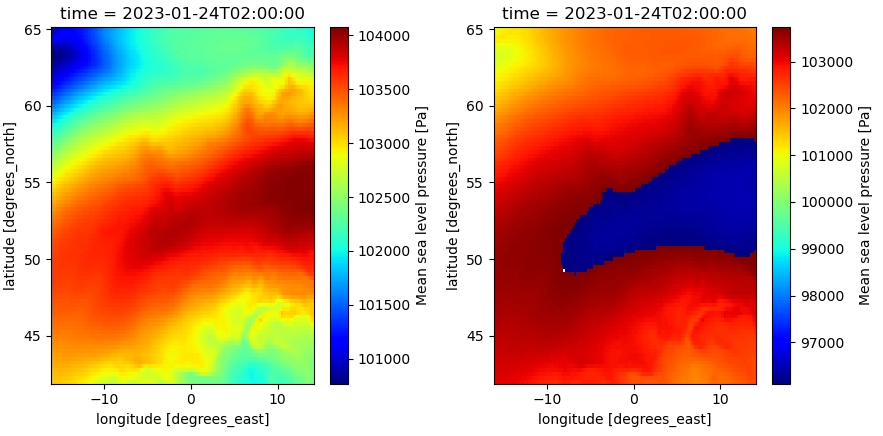
xarray.to_netcdf() of opened mfdataset results in incorrect data when not using manual encoding
This results in incorrect data in the written file (right):
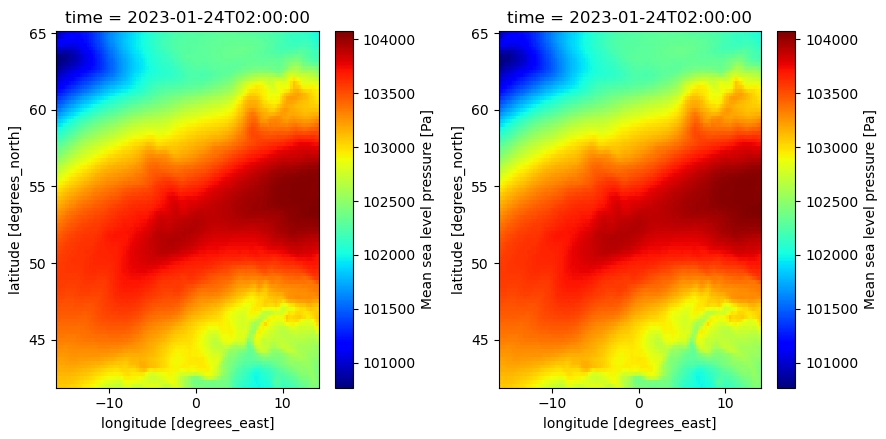
When updating the dtype (from int to float) in the variable encoding, this issues is solved:
The encoding in the source dataset:
Possible issue: source data is integers, but opening files with different scaling_factors (from different files) converts it to floats (or maybe this always happens). The dtype in the encoding is still int, so this is how the netcdf is written, but probably something does not fit within the int-bounds.
The text was updated successfully, but these errors were encountered: