This example shows how to create an ensemble of LAMMPS simulations using EasyVVUQ, execute the jobs through FabMD, then analyse them within the EasyVVUQ architecture. All within 3 FabSim commands!!!
Our tutorial uses the following components:
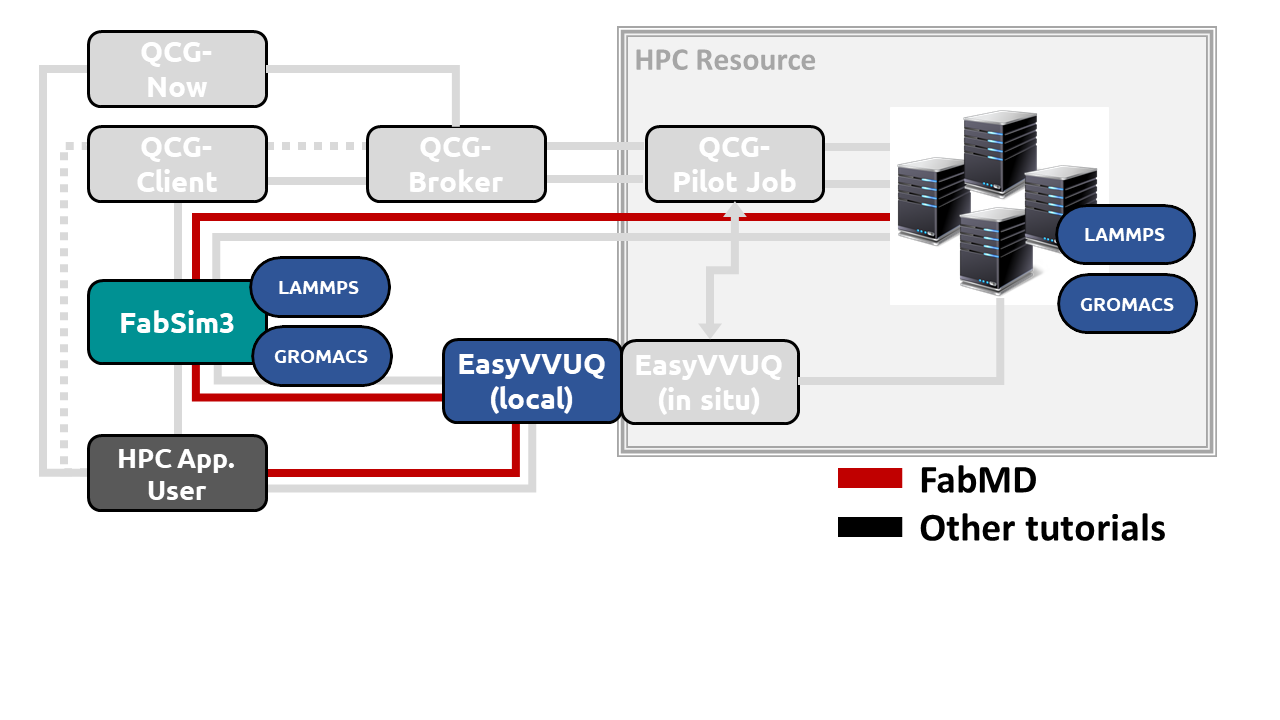
Its a very simple example of a LAMMPS ensemble. 5 replicas are created and given different velocity seeds. The solvation energy is calculated at the end of each simulation and the average and standard deviation are output to screen. The intention for this example is to provide a guide to designing your own workflow that uses these two tools together.
The input files needed for this example are found in plugins/FabMD/config_files/fabmd_easyvvuq. This directory contains three files:
-
lammps.template: is the LAMMPS input script, EasyVVUQ will substitute certain variables in this file to create the ensemble. -
data.peptide: is the configuration file for a peptide in water for LAMMPS simulation. This remains common to all simulations.
For this example it is assumed you can run the basic FabMD and EasyVVUQ examples.
These are the commands I needed to run this example:
fabsim archer easymd_example:fabmd_easyvvuq
fabsim archer job_stat # wait until execution has finished
fabsim archer easymd_example_analyse:fabmd_easyvvuq,fabmd_easyvvuq_archer_24
The generalised commands are:
fabsim remote_machine easymd_example:(name of config_dir in config_files)
fabsim remote_machine job_stat # wait until execution has finished
fabsim remote_machine easymd_example_analyse:(name of config_dir in config_files),(name of output_dir)
Each of the three commands is discussed in turn below. They are intended to serve as templates for your own workflows. See the commented code for more information in FabMD.py.
The first command easymd_example calls a function in FabMD.py. This sets up the ensemble with EasyVVUQ and executes them with FabSim on the remote machine.
EasyVVUQ requires a separate place to store the run information, this is put in a FabMD/tmp/.
An EasyVVUQ campagin is created in the usual way, we use a fixture to copy data.peptide to all directories.
Note that EasyVVUQ by default uses a $ to identify variables to carry out variable substitution, however $ is used by LAMMPS so we escape this clash by defining the delimiter to @.
We vary the random seed that LAMMPS uses to generate atomic velocities with: vary = {"velocity_seed": uq.distributions.uniform_integer(1,1000000)}
We sample 5 different velocity seeds and replicate each only once because given a specific seed the answer is deterministic. If you were varying a separate parameter that had a stochasitc ouput you would add replicas to each sample.
Then we save the campagin state for later, convert the campaign to a FabSim ensemble and execute a lammps_ensemble.
The second command is used to check when the simulatinos have completed. You could also use fabsim remote_machine stat.
The third command easymd_example_analyse calls a function in FabMD.py. This fetches the results, transfers it back to an EasyVVUQ campaign and then analyses it. It must be passed two arguments: the config_dir as usual; and the name of the results directory output_dir. output_dir will be the name of the directory containg the results in your local_results directory (set in your machines_user.yml file).
Note that we have made LAMMPS output a file that looks like a pandas dataframse csv file, a more general output could be read by EasyVVUQ by using a custom decoder.
This command does the equivalent of:
- fetching the results from the remote machine
- convert FabSim ensemble to EasyVVUQ campaign
- collate results into a pandas dataframe
- print all the information of from the campaign
- prints the
BasicStatson the solvation energy, including its mean and confidence intervals