|
| 1 | +{ |
| 2 | + "cells": [ |
| 3 | + { |
| 4 | + "cell_type": "markdown", |
| 5 | + "id": "01419663", |
| 6 | + "metadata": {}, |
| 7 | + "source": [ |
| 8 | + "# Matplotlib\n", |
| 9 | + "- A comprehensive library for creating static, animated, and interactive visualizations in Python.\n", |
| 10 | + "- Built on NumPy arrays and designed to work with the broader SciPy stack." |
| 11 | + ] |
| 12 | + }, |
| 13 | + { |
| 14 | + "cell_type": "code", |
| 15 | + "execution_count": null, |
| 16 | + "id": "0748b014", |
| 17 | + "metadata": {}, |
| 18 | + "outputs": [], |
| 19 | + "source": [ |
| 20 | + "import matplotlib as mpl\n", |
| 21 | + "import matplotlib.pyplot as plt\n", |
| 22 | + "import numpy as np" |
| 23 | + ] |
| 24 | + }, |
| 25 | + { |
| 26 | + "cell_type": "markdown", |
| 27 | + "id": "348c1b11", |
| 28 | + "metadata": {}, |
| 29 | + "source": [ |
| 30 | + "## Basic Line Plot\n", |
| 31 | + "From Getting Started." |
| 32 | + ] |
| 33 | + }, |
| 34 | + { |
| 35 | + "cell_type": "code", |
| 36 | + "execution_count": null, |
| 37 | + "id": "37ad8b55", |
| 38 | + "metadata": {}, |
| 39 | + "outputs": [], |
| 40 | + "source": [ |
| 41 | + "fig, ax = plt.subplots() # Create a figure containing a single Axes.\n", |
| 42 | + "ax.plot([1, 2, 3, 4], [1, 4, 2, 3]) # Plot some data on the Axes.\n", |
| 43 | + "plt.show() # Show the figure." |
| 44 | + ] |
| 45 | + }, |
| 46 | + { |
| 47 | + "cell_type": "markdown", |
| 48 | + "id": "dd74e2cc", |
| 49 | + "metadata": { |
| 50 | + "lines_to_next_cell": 0 |
| 51 | + }, |
| 52 | + "source": [ |
| 53 | + "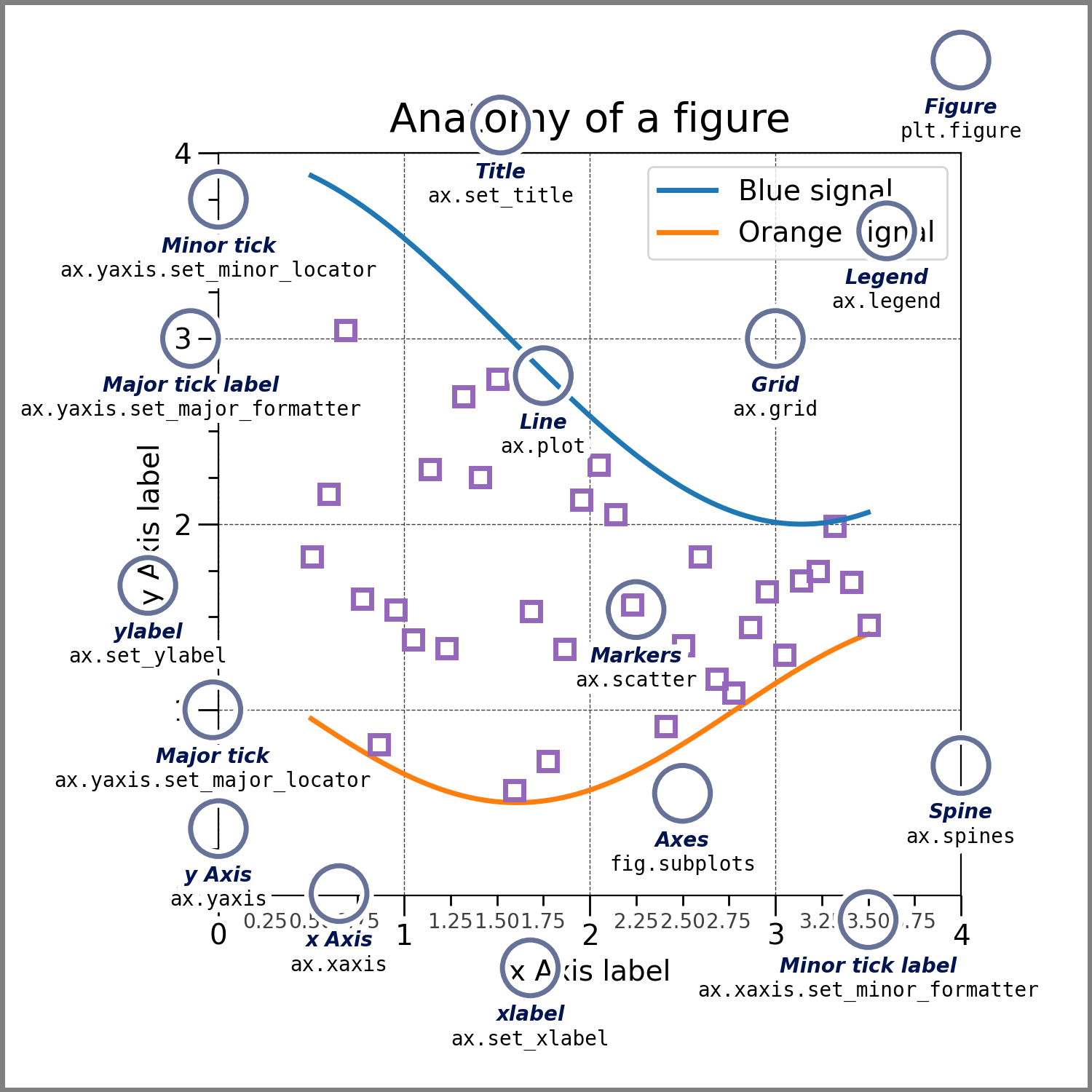" |
| 54 | + ] |
| 55 | + }, |
| 56 | + { |
| 57 | + "cell_type": "code", |
| 58 | + "execution_count": null, |
| 59 | + "id": "2d86fcd1", |
| 60 | + "metadata": {}, |
| 61 | + "outputs": [], |
| 62 | + "source": [ |
| 63 | + "fig, ax = plt.subplots() # Create a figure containing a single Axes.\n", |
| 64 | + "ax.plot([1, 2, 3, 4], [1, 4, 2, 3]) # Plot some data on the Axes.\n", |
| 65 | + "ax.grid()\n", |
| 66 | + "plt.show()" |
| 67 | + ] |
| 68 | + }, |
| 69 | + { |
| 70 | + "cell_type": "markdown", |
| 71 | + "id": "0595fdfd", |
| 72 | + "metadata": {}, |
| 73 | + "source": [ |
| 74 | + "Customize ticks\n", |
| 75 | + "- [Ticker API](https://matplotlib.org/stable/api/ticker_api.html)" |
| 76 | + ] |
| 77 | + }, |
| 78 | + { |
| 79 | + "cell_type": "code", |
| 80 | + "execution_count": null, |
| 81 | + "id": "0817796f", |
| 82 | + "metadata": {}, |
| 83 | + "outputs": [], |
| 84 | + "source": [ |
| 85 | + "ax.yaxis.set_major_locator(mpl.ticker.MaxNLocator(integer=True))\n", |
| 86 | + "# ax.xaxis.set_major_locator(mpl.ticker.MaxNLocator(integer=True))\n", |
| 87 | + "ax.get_figure()" |
| 88 | + ] |
| 89 | + }, |
| 90 | + { |
| 91 | + "cell_type": "code", |
| 92 | + "execution_count": null, |
| 93 | + "id": "80d2be97", |
| 94 | + "metadata": {}, |
| 95 | + "outputs": [], |
| 96 | + "source": [ |
| 97 | + "mu, sigma = 115, 15\n", |
| 98 | + "x = mu + sigma * np.random.randn(10000)\n", |
| 99 | + "fig, ax = plt.subplots(figsize=(5, 2.7), layout=\"constrained\")\n", |
| 100 | + "# the histogram of the data\n", |
| 101 | + "n, bins, patches = ax.hist(x, 50, density=True, facecolor=\"C0\", alpha=0.75)\n", |
| 102 | + "\n", |
| 103 | + "ax.set_xlabel(\"Length [cm]\")\n", |
| 104 | + "ax.set_ylabel(\"Probability\")\n", |
| 105 | + "ax.set_title(\"Aardvark lengths\\n (not really)\")\n", |
| 106 | + "ax.text(75, 0.025, r\"$\\mu=115,\\ \\sigma=15$\")\n", |
| 107 | + "ax.axis([55, 175, 0, 0.03])\n", |
| 108 | + "ax.grid(True)" |
| 109 | + ] |
| 110 | + }, |
| 111 | + { |
| 112 | + "cell_type": "code", |
| 113 | + "execution_count": null, |
| 114 | + "id": "d78dc9ce", |
| 115 | + "metadata": {}, |
| 116 | + "outputs": [], |
| 117 | + "source": [ |
| 118 | + "# ## Proteomics data example" |
| 119 | + ] |
| 120 | + }, |
| 121 | + { |
| 122 | + "cell_type": "code", |
| 123 | + "execution_count": null, |
| 124 | + "id": "c290244b", |
| 125 | + "metadata": {}, |
| 126 | + "outputs": [], |
| 127 | + "source": [ |
| 128 | + "import pathlib\n", |
| 129 | + "\n", |
| 130 | + "import pandas as pd\n", |
| 131 | + "\n", |
| 132 | + "dir_data = pathlib.Path(\"data\")\n", |
| 133 | + "df = pd.read_csv(dir_data / \"proteins\" / \"proteins.csv\", index_col=0)\n", |
| 134 | + "df" |
| 135 | + ] |
| 136 | + }, |
| 137 | + { |
| 138 | + "cell_type": "code", |
| 139 | + "execution_count": null, |
| 140 | + "id": "1787a67d", |
| 141 | + "metadata": {}, |
| 142 | + "outputs": [], |
| 143 | + "source": [ |
| 144 | + "x = df.iloc[0]\n", |
| 145 | + "x" |
| 146 | + ] |
| 147 | + }, |
| 148 | + { |
| 149 | + "cell_type": "code", |
| 150 | + "execution_count": null, |
| 151 | + "id": "8e27f422", |
| 152 | + "metadata": {}, |
| 153 | + "outputs": [], |
| 154 | + "source": [ |
| 155 | + "ax = x.hist()" |
| 156 | + ] |
| 157 | + }, |
| 158 | + { |
| 159 | + "cell_type": "code", |
| 160 | + "execution_count": null, |
| 161 | + "id": "5e525e25", |
| 162 | + "metadata": { |
| 163 | + "lines_to_next_cell": 2 |
| 164 | + }, |
| 165 | + "outputs": [], |
| 166 | + "source": [ |
| 167 | + "fig, ax = plt.subplots()\n", |
| 168 | + "n, bins, patches = ax.hist(x, bins=30, alpha=0.7, color=\"C0\")" |
| 169 | + ] |
| 170 | + }, |
| 171 | + { |
| 172 | + "cell_type": "markdown", |
| 173 | + "id": "6360d4cd", |
| 174 | + "metadata": {}, |
| 175 | + "source": [ |
| 176 | + "## Available styles\n", |
| 177 | + "Choose your preferred style with it's defaults\n", |
| 178 | + "[here](https://matplotlib.org/stable/gallery/style_sheets/style_sheets_reference.html)\n", |
| 179 | + "\n", |
| 180 | + "```python\n", |
| 181 | + "plt.style.use('ggplot')\n", |
| 182 | + "```\n", |
| 183 | + "\n", |
| 184 | + "\n", |
| 185 | + "\n", |
| 186 | + "" |
| 187 | + ] |
| 188 | + }, |
| 189 | + { |
| 190 | + "cell_type": "code", |
| 191 | + "execution_count": null, |
| 192 | + "id": "f32690b4", |
| 193 | + "metadata": {}, |
| 194 | + "outputs": [], |
| 195 | + "source": [ |
| 196 | + "with plt.style.context(\"ggplot\"):\n", |
| 197 | + " fig, ax = plt.subplots()\n", |
| 198 | + " n, bins, patches = ax.hist(x, bins=30, alpha=0.7)" |
| 199 | + ] |
| 200 | + }, |
| 201 | + { |
| 202 | + "cell_type": "code", |
| 203 | + "execution_count": null, |
| 204 | + "id": "41efb93a", |
| 205 | + "metadata": {}, |
| 206 | + "outputs": [], |
| 207 | + "source": [] |
| 208 | + } |
| 209 | + ], |
| 210 | + "metadata": { |
| 211 | + "jupytext": { |
| 212 | + "cell_metadata_filter": "-all", |
| 213 | + "main_language": "python", |
| 214 | + "notebook_metadata_filter": "-all" |
| 215 | + } |
| 216 | + }, |
| 217 | + "nbformat": 4, |
| 218 | + "nbformat_minor": 5 |
| 219 | +} |
0 commit comments