|
| 1 | +# Tips for spatiotemporal indexing |
| 2 | + |
| 3 | + |
| 4 | +In the both the [AdaSTEM Demo](https://chenyangkang.github.io/stemflow/Examples/01.AdaSTEM_demo.html) and [SphereAdaSTEM demo](https://chenyangkang.github.io/stemflow/Examples/04.SphereAdaSTEM_demo.html) we use bird observation data to demonstrate functionality of AdaSTEM. Spatiotemporal coordinate are homogeneously encoded in these two cases, with `longitude` and `latitude` being spatial indexes and `DOY` (day of year) being temporal index. |
| 5 | + |
| 6 | +Here, we present more tips and examples on how to play with these indexing systems. |
| 7 | + |
| 8 | +------ |
| 9 | + |
| 10 | + |
| 11 | +## 2D + Temporal indexing |
| 12 | + |
| 13 | +### Flexible coordinate systems |
| 14 | + |
| 15 | +`stemflow` support all types of spatial coordinate reference system (CRS) and temporal indexing (for example, week month, year, or decades). `stemflow` only support tabular point data currently. You should transform your data to desired CRS before feeding them to `stemflow`. |
| 16 | + |
| 17 | +For example, transforming CRS: |
| 18 | + |
| 19 | +```python |
| 20 | +import pyproj |
| 21 | + |
| 22 | +# Define the source and destination coordinate systems |
| 23 | +source_crs = pyproj.CRS.from_epsg(4326) # WGS 84 (latitude, longitude) |
| 24 | +target_crs = pyproj.CRS.from_string("ESRI:54017") # World Behrmann equal area projection (x, y) |
| 25 | + |
| 26 | +# Create a transformer object |
| 27 | +transformer = pyproj.Transformer.from_crs(source_crs, target_crs, always_xy=True) |
| 28 | + |
| 29 | +# Project |
| 30 | +data['proj_lng'], data['proj_lat'] = transformer.transform(data['lng'].values, data['lat'].values) |
| 31 | +``` |
| 32 | + |
| 33 | +Now the projected spatial coordinate for each record is stored in `data['proj_lng']` and `data['proj_lat']` |
| 34 | +We can then feed this data to `stemflow`: |
| 35 | + |
| 36 | + |
| 37 | + |
| 38 | + |
| 39 | +```python |
| 40 | + |
| 41 | +from stemflow.model.AdaSTEM import AdaSTEMClassifier |
| 42 | +from xgboost import XGBClassifier |
| 43 | + |
| 44 | +model = AdaSTEMClassifier( |
| 45 | + base_model=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0,n_jobs=1), |
| 46 | + save_gridding_plot = True, |
| 47 | + ensemble_fold=10, # data are modeled 10 times, each time with jitter and rotation in Quadtree algo |
| 48 | + min_ensemble_required=7, # Only points covered by > 7 stixels will be predicted |
| 49 | + grid_len_upper_threshold=1e5, # force splitting if the edge of grid exceeds 1e5 meters |
| 50 | + grid_len_lower_threshold=1e3, # stop splitting if the edge of grid fall short 1e3 meters |
| 51 | + temporal_start=1, # The next 4 params define the temporal sliding window |
| 52 | + temporal_end=52, |
| 53 | + temporal_step=2, |
| 54 | + temporal_bin_interval=4, |
| 55 | + points_lower_threshold=50, # Only stixels with more than 50 samples are trained |
| 56 | + Spatio1='proj_lng', # Use the column 'proj_lng' and 'proj_lat' as spatial indexes |
| 57 | + Spatio2='proj_lat', |
| 58 | + Temporal1='Week', |
| 59 | + use_temporal_to_train=True, # In each stixel, whether 'Week' should be a predictor |
| 60 | + njobs=1 |
| 61 | +) |
| 62 | +``` |
| 63 | + |
| 64 | +Here, we use temporal bin of 4 weeks and step of 2 weeks, starting from week 1 to week 52. For spatial indexing, we force the gird size to be `1km (1e3 m) ~ 10km (1e5 m)`. Since `ESRI 54017` is an equal area projection, the unit is meter. |
| 65 | + |
| 66 | + |
| 67 | +Then we could fit the model: |
| 68 | + |
| 69 | +```py |
| 70 | +## fit |
| 71 | +model = model.fit(data.drop('target', axis=1), data[['target']]) |
| 72 | + |
| 73 | +## predict |
| 74 | +pred = model.predict(X_test) |
| 75 | +pred = np.where(pred<0, 0, pred) |
| 76 | +eval_metrics = AdaSTEM.eval_STEM_res('classification',y_test, pred_mean) |
| 77 | +``` |
| 78 | + |
| 79 | +Note that the [Quadtree [1]](https://dl.acm.org/doi/abs/10.1145/356924.356930) algo is limited to 6 digits for efficiency. So transform your coordinate of it exceeds that threshold. For example, x=0.0000001 and y=0.0000012 will be problematic. Consider changing them to x=100 and y=1200. |
| 80 | + |
| 81 | +------ |
| 82 | +### Spatial-only modeling |
| 83 | + |
| 84 | +By playing some tricks, you can also do a `spatial-only` modeling, without splitting the data into temporal blocks: |
| 85 | + |
| 86 | +```python |
| 87 | +model = AdaSTEMClassifier( |
| 88 | + base_model=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0,n_jobs=1), |
| 89 | + save_gridding_plot = True, |
| 90 | + ensemble_fold=10, |
| 91 | + min_ensemble_required=7, |
| 92 | + grid_len_upper_threshold=1e5, |
| 93 | + grid_len_lower_threshold=1e3, |
| 94 | + temporal_start=1, |
| 95 | + temporal_end=52, |
| 96 | + temporal_step=1000, # Setting step and interval largely outweigh |
| 97 | + temporal_bin_interval=1000, # temporal scale of data |
| 98 | + points_lower_threshold=50, |
| 99 | + Spatio1='proj_lng', |
| 100 | + Spatio2='proj_lat', |
| 101 | + Temporal1='Week', |
| 102 | + use_temporal_to_train=True, |
| 103 | + njobs=1 |
| 104 | +) |
| 105 | +``` |
| 106 | + |
| 107 | +Setting `temporal_step` and `temporal_bin_interval` largely outweigh the temporal scale (1000 compared with 52) of your data will render only `one` temporal window during splitting. Consequently, your model would become a spatial model. This could be beneficial if temporal heterogeneity is not of interest, or without enough data to investigate. |
| 108 | + |
| 109 | +------- |
| 110 | + |
| 111 | +### Fix the gird size of Quadtree algorithm |
| 112 | + |
| 113 | +There are **two ways** to fix the grid size: |
| 114 | + |
| 115 | +#### 1. By using some tricks we can fix the gird size/edge length of AdaSTEM model classes: |
| 116 | + |
| 117 | +```python |
| 118 | +model = AdaSTEMClassifier( |
| 119 | + base_model=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0,n_jobs=1), |
| 120 | + save_gridding_plot = True, |
| 121 | + ensemble_fold=10, |
| 122 | + min_ensemble_required=7, |
| 123 | + grid_len_upper_threshold=1000, |
| 124 | + grid_len_lower_threshold=1000, |
| 125 | + temporal_start=1, |
| 126 | + temporal_end=52, |
| 127 | + temporal_step=2, |
| 128 | + temporal_bin_interval=4, |
| 129 | + points_lower_threshold=0, |
| 130 | + stixel_training_size_threshold=50, |
| 131 | + Spatio1='proj_lng', |
| 132 | + Spatio2='proj_lat', |
| 133 | + Temporal1='Week', |
| 134 | + use_temporal_to_train=True, |
| 135 | + njobs=1 |
| 136 | +) |
| 137 | +``` |
| 138 | + |
| 139 | +Quadtree will keep splitting until it hits an edge length lower than 1000 meters. Data volume won't hamper this process because the splitting threshold is set to 0 (`points_lower_threshold=0`). Stixels with sample volume less than 50 still won't be trained (`stixel_training_size_threshold=50`). However, we cannot guarantee the exact grid length. It should be somewhere between 500m and 1000m since each time Quadtree do a bifurcated splitting. |
| 140 | + |
| 141 | +#### 2. Using `STEM` model classes |
| 142 | + |
| 143 | +We also implemented `STEM` model classes for fixed gridding. Instead of adaptive splitting based on data abundance, `STEM` model classes split the space with fixed grid length: |
| 144 | + |
| 145 | +```python |
| 146 | +from stemflow.model.STEM import STEM, STEMRegressor, STEMClassifier |
| 147 | + |
| 148 | +model = STEMClassifier( |
| 149 | + base_model=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0,n_jobs=1), |
| 150 | + save_gridding_plot = True, |
| 151 | + ensemble_fold=10, |
| 152 | + min_ensemble_required=7, |
| 153 | + grid_len=1000, |
| 154 | + temporal_start=1, |
| 155 | + temporal_end=52, |
| 156 | + temporal_step=2, |
| 157 | + temporal_bin_interval=4, |
| 158 | + points_lower_threshold=0, |
| 159 | + stixel_training_size_threshold=50, |
| 160 | + Spatio1='proj_lng', |
| 161 | + Spatio2='proj_lat', |
| 162 | + Temporal1='Week', |
| 163 | + use_temporal_to_train=True, |
| 164 | + njobs=1 |
| 165 | +) |
| 166 | +``` |
| 167 | + |
| 168 | +Here, `grid_len` parameter take place the original upper and lower threshold parameters. The main functionality is the same as `AdaSTEM` classes. |
| 169 | + |
| 170 | +---- |
| 171 | +## 3D spherical + Temporal indexing |
| 172 | + |
| 173 | +Our earth is a sphere, and consequently there is no single solution to project the sphere to a 2D plane while maintaining the distance and area – all projection method as pros and cons. We also implemented spherical indexing to solve this issue. |
| 174 | + |
| 175 | + |
| 176 | +```python |
| 177 | +from stemflow.model.SphereAdaSTEM import SphereAdaSTEMRegressor |
| 178 | +from xgboost import XGBClassifier, XGBRegressor |
| 179 | + |
| 180 | +model = SphereAdaSTEMRegressor( |
| 181 | + base_model=Hurdle( |
| 182 | + classifier=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1), |
| 183 | + regressor=XGBRegressor(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1) |
| 184 | + ), # hurdel model for zero-inflated problem (e.g., count) |
| 185 | + save_gridding_plot = True, |
| 186 | + ensemble_fold=10, # data are modeled 10 times, each time with jitter and rotation in Quadtree algo |
| 187 | + min_ensemble_required=7, # Only points covered by > 7 stixels will be predicted |
| 188 | + grid_len_upper_threshold=2500, # force splitting if the grid length exceeds 2500 (km) |
| 189 | + grid_len_lower_threshold=500, # stop splitting if the grid length fall short 500 (km) |
| 190 | + temporal_start=1, # The next 4 params define the temporal sliding window |
| 191 | + temporal_end=366, |
| 192 | + temporal_step=25, # The window takes steps of 20 DOY (see AdaSTEM demo for details) |
| 193 | + temporal_bin_interval=50, # Each window will contain data of 50 DOY |
| 194 | + points_lower_threshold=50, # Only stixels with more than 50 samples are trained |
| 195 | + Temporal1='DOY', |
| 196 | + use_temporal_to_train=True, # In each stixel, whether 'DOY' should be a predictor |
| 197 | + njobs=1 |
| 198 | +) |
| 199 | +``` |
| 200 | + |
| 201 | +`SphereAdaSTEM` module has almost the same structure and functions as `AdaSTEM` and `STEM` modules. The only difference is that |
| 202 | + |
| 203 | +1. It mandatorily looks for "longitude" and "latitude" in the columns. |
| 204 | +1. It splits the data using [`Sphere QuadTree` [2]](https://ieeexplore.ieee.org/abstract/document/146380). |
| 205 | +1. It plots the grids using `plotly`. |
| 206 | + |
| 207 | + |
| 208 | +See [SphereAdaSTEM demo](https://chenyangkang.github.io/stemflow/Examples/04.SphereAdaSTEM_demo.html) and [Interactive spherical gridding plot](https://chenyangkang.github.io/stemflow/assets/Sphere_gridding.html). |
| 209 | + |
| 210 | +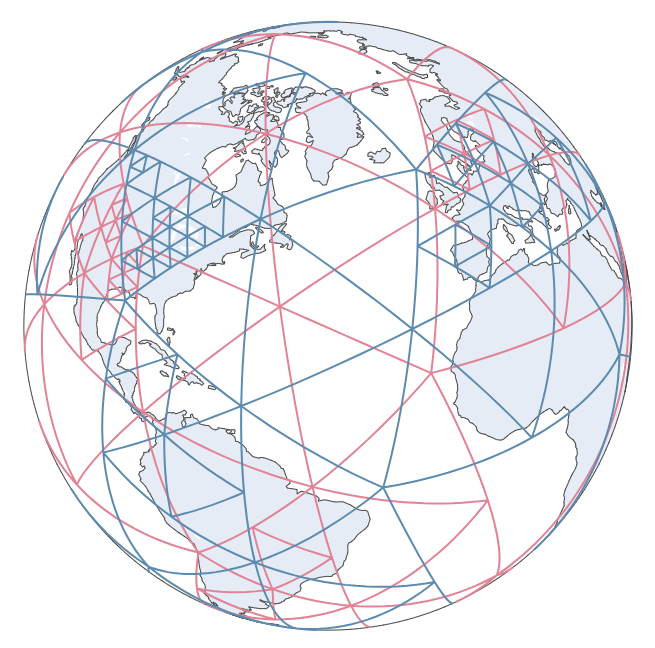 |
| 211 | + |
| 212 | +----- |
| 213 | +## References: |
| 214 | + |
| 215 | +1. [Samet, H. (1984). The quadtree and related hierarchical data structures. ACM Computing Surveys (CSUR), 16(2), 187-260.](https://dl.acm.org/doi/abs/10.1145/356924.356930) |
| 216 | + |
| 217 | +1. [Gyorgy, F. (1990, October). Rendering and managing spherical data with sphere quadtrees. In Proceedings of the First IEEE Conference on Visualization: Visualization90 (pp. 176-186). IEEE.](https://ieeexplore.ieee.org/abstract/document/146380) |
0 commit comments