Shift in energy test set and pressure #2935
Replies: 9 comments
-
|
Do your two datasets have the same chemical composition? |
Beta Was this translation helpful? Give feedback.
-
|
Yeah, it is NH3 |
Beta Was this translation helpful? Give feedback.
-
|
The shift in virial is not expected. Could you please check, by your labeling method, if the virial is equal to the derivative of the energy? |
Beta Was this translation helpful? Give feedback.
-
|
Do you mean that you want me to check if the first-principles calculations (made with quantum espresso) are correct? I've already checked the convergence of the viral tensor and it's converged with respect to the plane waves cut off that I used. I've also noticed that training on the virial seems to reduce this shift but I was wondering why is it a rigid shift and not just a random error of the NN. It seems like there is some kind of bias somewhere. |
Beta Was this translation helpful? Give feedback.
-
|
You may check if the following equation holds where |
Beta Was this translation helpful? Give feedback.
-
|
Can you provide an example for reproducing? |
Beta Was this translation helpful? Give feedback.
-
|
Here's the link
|
Beta Was this translation helpful? Give feedback.
-
|
I plot In this figure, the bias is about 5 eV for 576 atoms instead of 15 eV in your figure. Note that your training data has a similar bias. |
Beta Was this translation helpful? Give feedback.
-
|
Yes, you are right, the model that I linked you is trained over 2 million steps while the one of the plots over 1 million steps. I LINK you the correct model.
You can see that theres is a shift both in energy and virial (diagonal) in the test_data while there's not in the train_data. Is this only a matter of the quality of the training, in the sense that increasing the number of training steps would solve the problem? Why is the error such a rigid shift in the diagonal terms of the virial? Why are the off-diagonal element of the virial instead correct? |
Beta Was this translation helpful? Give feedback.

-
Summary
Shift in energy test set and pressure when I run it on a dataset with different number of atoms with respect to the training one.
DeePMD-kit Version
v2.1.1.dev80+g8c58455
TensorFlow Version
2.3.0
Python Version, CUDA Version, GCC Version, LAMMPS Version, etc
No response
Details
Hi, I trained a NN over a data set made of snapshots a 432 atoms system. When I test this NN over a different dataset made of 576 atoms ( of course not included in the train one) I find good predictions for forces, so the dynamics of the system should be fine, but I find a rigid shift upon the total energy and also on the diagonal elements of the viral.
energy
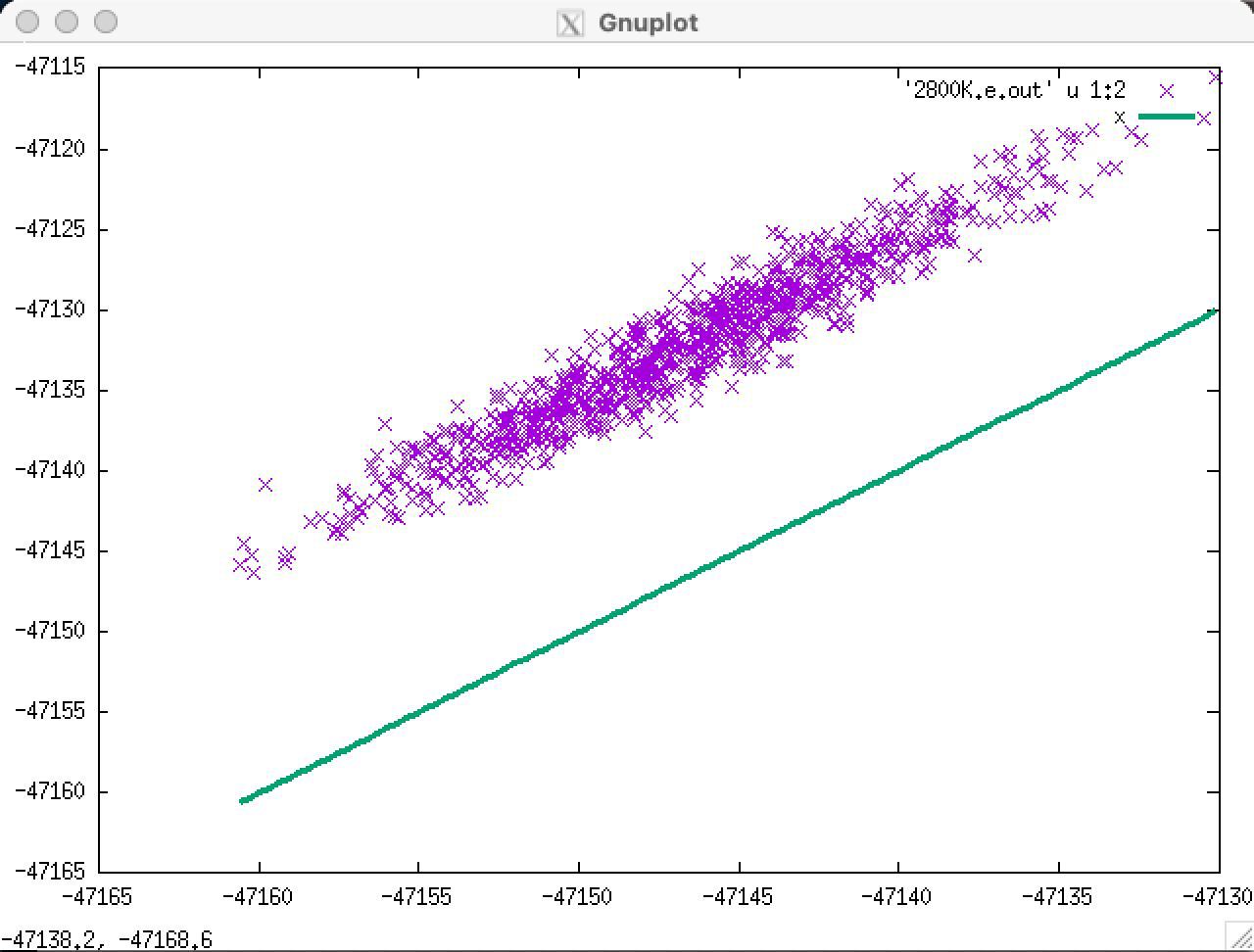
yy diagonal element of the virial

On the training set these shifts are not present. I thought that specifying the atomic energies of each species would have solve the problem, but It did not. Is there any kind of solution to this problem?
Thanks
Beta Was this translation helpful? Give feedback.
All reactions