-
Notifications
You must be signed in to change notification settings - Fork 14
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
rendering tab characters in .Rmd #9
Comments
|
The htmlTable() is now in a separate package called... htmlTable! It has been submitted to CRAN and should be available within short. Once it is published the Gmisc package will depend on htmlTable for that part of functionality. It seems that the first examples contain valid code but that the print.htmlTable fails to use the viewer. I can't reproduce the first error using the htmlTable-package, could you try that package and see if you still get the error: |
|
I still just get a pile of html code, but library(htmlTable)
tmp <- htmlTable(mtcars)
gsub('\t','', tmp)works as does "test.Rmd" {r, results='asis'}
library(htmlTable)
htmlTable(mtcars)
Bizarre.. but at least I have a couple work-arounds. (I'm running all the updated packages and rstudio and 3.1.2) |
|
I can't reproduce the bug :-( I've added a test-case that should reproduce your issue - try to run it and see if you still get the same error issues. It sounds like it could possibly be an issue related to iOS/Linux versions of RStudio/rmarkdown. You can also try and force the non-interactive mode by |
|
I recently got a bug report from another user with the same problem and it turned out that it was related to a bug in his |
|
I still don't know.. It is definitely a pandoc issue. Possibly more of a pandoc/mac issue. I had already tried reinstalling it (using a package and from source), and that didn't work. Finally, I went back to 1.12.3 (in hindsight, I suppose this started after I updated to 1.13 which has many warnings about behavioral changes that may break existing documents). Maybe I will look into this again when I don't have this pandoc migraine and some more free time. So everything seems to be working as expected, but when rendering, now rstudio uses Do you know what platform the other user with this problem was using and which version of pandoc? |
|
It seems that the 1.13 was causing his issue as well, although he is using Windows. You may want to write a bug report to pandoc: https://github.com/jgm/pandoc/issues |
|
The bug comes from version 1.13.1. I am using both Windows 8.1 and also 7.1 pandoc 1.13.1 My ``quick'' fix on my side was to rename Pandoc folder to OldPandoc and everything revert back to normal as long as knitr was calling pandoc from within Rstudio/bin/pandoc/pandoc. |
|
Since it will most likely take a while before the pandoc issue is properly addressed, could you check if the bugfix in the develop branch works: |
|
It did not work on my side. I think the quick fix is the best bet for now. It does not change any of the knitr functionality etc (at least on my end). |
|
Ok, I think I've nailed it - pandoc (correctly) classifies the indentation as code blocks. There is a bug report here discussing possible solutions. Until we have a better solution I have fixed the issue by removing all the indentation in the |
|
This does fix the problem in 1.13 using the develop branch, but I agree that the readability of the non-indented code suffers greatly. If/when the pandoc issue is resolved, it should not affect the functionality of your package, and it would be easy to add the indentation for aesthetic purposes as a patch. Thanks again. |

I've never had problems rendering tables in markdown documents until recently, and I can't quite figure out if this is happening due to a knitr, rmarkdown, or R3.1.2 update. Do you see anything like this as well?
If I have a .Rmd with
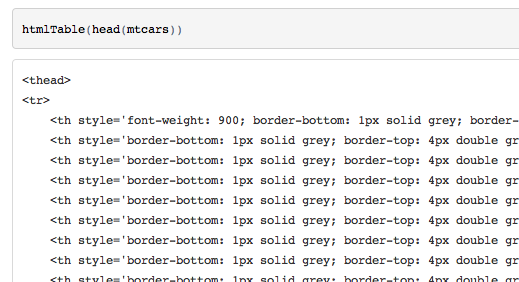
{r, results='asis'} library("Gmisc") htmlTable(head(mtcars))I get the results
current cran version
github
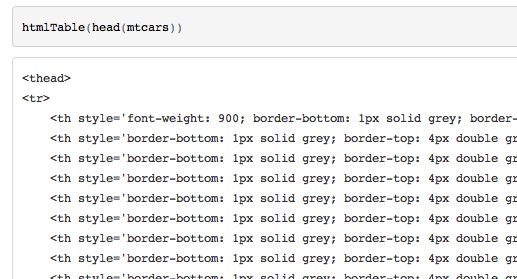
github v1.0
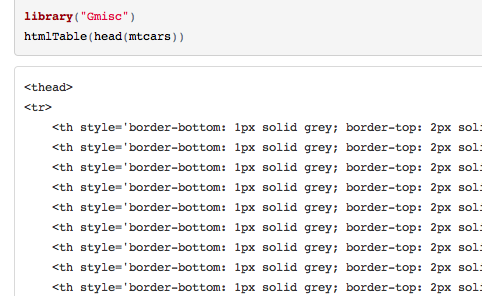
Subbing out all the tab characters in
table_strrenders it properlygsub("\t", " ", x)but it wasn't an issue before. Can you reproduce this?
The text was updated successfully, but these errors were encountered: