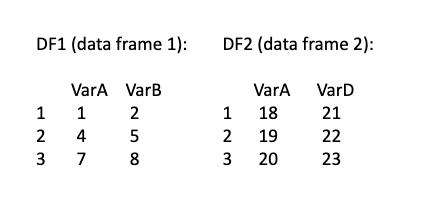
- A matrix may contain numeric values only.
- A matrix must not be singular.
- A data frame may contain variables that have different modes.
- A data frame may contain variables of different lengths.
unclass(as.Date("1971-01-01"))
- 1
- 365
- 4
- 12
- remove()
- erase()
- detach()
- delete()
xvect<-c(1,2,3)
xvect[2] <- "2"
xvect
- [1] 1 2 3
- [1] "1" 2 "3"
- [1] "1" "2" "3"
- [1] 7 9
Q5. The variable height is a numeric vector in the code below. Which statement returns the value 35?
-
height(length(height)) -
height[length(height)] -
height[length[height]] -
height(5)
Q6. In the image below, the data frame is named rates. The statement sd(rates[, 2]) returns 39. As what does R regard Ellen's product ratings?
- sample with replacement
- population
- trimmed sample
- sample <-- not sure
-
Var_A! -
\_VarA -
.2Var_A -
Var2_A
- A matrix has two dimensions, while an array can have three or more dimensions.
- An array is a subtype of the data frame, while a matrix is a separate type entirely.
- A matrix can have columns of different lengths, but an array's columns must all be the same length.
- A matrix may contain numeric values only, while an array can mix different types of values.
- type
- length
- attributes
- scalar
Q10. In the image below, the data frame on lines 1 through 4 is named StDf. State and Capital are both factors. Which statement returns the results shown on lines 6 and 7?
- StDf[1:2,-3]
- StDf[1:2,1]
- StDf[1:2,]
- StDf[1,2,]
- BOF(pizza, 5)
- first(pizza, 5)
- top(pizza, 5)
- head(pizza, 5)
Q12. You accidentally display a large data frame on the R console, losing all the statements you entered during the current session. What is the best way to get the prior 25 statements back?
- console(-25)
- console(reverse=TRUE)
- history()
- history(max.show = 25)
Q13. d.pizza is a data frame. It's a column named temperature contains only numbers. If you extract temperature using the [] accessors, its class defaults to numeric. How can you access temperature so that it retains the class of data.frame?
> class( d.pizza[ , "temperature" ] )
> "numeric"
-
class( d.pizza( , "temperature" ) ) -
class( d.pizza[ , "temperature" ] ) -
class( d.pizza$temperature ) -
class( d.pizza[ , "temperature", drop=F ] )
a <- c(3,3,6.5,8)
b <- c(7,2,5.5,10)
c <- a < b
- [1] NaN
- [1] -4
- [1] 4 -1 -1 2
- [1] TRUE FALSE FALSE TRUE
Q15. Review the statements below. Does the use of the dim function change the class of y, and if so what is y's new class?
> y <- 1:9
> dim(y) <- c(3,3)
- No, y's new class is "array".
- Yes, y's new class is "matrix".
- No, y's new class is "vector".
- Yes, y's new class is "integer".
mydf <- data.frame(x=1:3, y=c("a","b","c"), stringAsFactors=FALSE)
- list
- string
- factor
- character vector
- Vectors are used only for numeric data, while lists are useful for both numeric and string data.
- Vectors and lists are the same thing and can be used interchangeably.
- A vector contains items of a single data type, while a list can contain items of different data types.
- Vectors are like arrays, while lists are like data frames.
- list.objects()
- print.objects()
- getws()
- ls()
- rbind()
- cbind()
- bind()
- coerce()
1 mylist <- list(1,2,"C",4,5)
2 unlist(mylist)
- [1] 1 2 4 5
- "C"
- [1] "1" "2" "C" "4" "5"
- [1] 1 2 C 4 5
x <- NA
y <- x/1
- Inf
- Null
- NaN
- NA
Q22. Two variable in the mydata data frame are named Var1 and Var2. How do you tell a bivariate function, such as cor.test, which two variables you want to analyze?
-
cor.test(Var1 ~ Var2) -
cor.test(mydata$(Var1,Var2)) -
cor.test(mydata$Var1,mydata$Var2) -
cor.test(Var1,Var2, mydata)
Q23. A data frame named d.pizza is part of the DescTools package. A statement is missing from the following R code and an error is therefore likely to occur. Which statement is missing?
library(DescTools)
deliver <- aggregate(count,by=list(area,driver), FUN=mean)
print(deliver)
-
attach(d.pizza) -
summarize(deliver) -
mean <- rbind(d.pizza,count) -
deliver[!complete.cases(deliver),]
- data frame: names() and rownames() matrix: colnames() and row.names()
- data frame: names() and row.names() matrix: dimnames() (not sure)
- data frame: colnames() and row.names() matrix: names() and rownames()
- data frame: colnames() and rownames() matrix: names() and row.names()
Q25. Which set of two statements-followed by the cbind() function-results in a data frame named vbound?
- [ ]
v1<-list(1,2,3)
v2<-list(c(4,5,6))
vbound<-cbind(v1,v2)
- [ ]
v1<-c(1,2,3)
v2<-list(4,5,6))
vbound<-cbind(v1,v2)
- [ ]
v1<-c(1,2,3)
v2<-c(4,5,6))
vbound<-cbind(v1,v2)
Cpeople <- ournames %in% grep("^C", ournames, value=TRUE)
- records where the first character is a C
- any record with a value containing a C
- TRUE or FALSE, depending on whether any character in ournames is C
- TRUE and FALSE values, depending on whether the first character in an ournames record is C
v <- 1:3
names(v) <- c("a", "b", "c")
v[4] <- 4
- ""
- d
- NULL
- NA
Q28. Which of the following statements doesn't yield the code output below. Review the following code. What is the result of line 3?
x <- c(1, 2, 3, 4)
Output: [1] 2 3 4
- x[c(2, 3, 4)]
- x[-1]
- x[c(-1, 0, 0, 0)]
- x[c(-1, 2, 3, 4)]
- 6
- 9
- 3
- 0
x<-5:8
names(x)<-letters[5:8]
x
- e f g h "5" "6" "7" "8"
- 5 6 7 8
- e f g h
- e f g h 5 6 7 8
x<-as.Date("2018-10-01")
- attr()
- months(x)
- as.month(x)
- month(x)
fact<-factor(c("Rep","Dem","Dem","Rep"))
fact
[1] Rep Dem Dem Rep
Levels: Rep Dem
fact[2]<-"Ind"
- >
- [,2]Ind
- invalid factor level, NA generated
- Ind
StartDate<- as.Date("2020/2/28")
StopDate<- as.Date("2020/3/1")
StopDate-StartDate
- "1970-01-02"
- time difference of one day
- time difference of two days
- error in x-y: nonnumeric argument to binary operator
> mtrx <- matrix( c(3,5,8,4), nrow= 2,ncol=2,byrow=TRUE)
> newmat <- mtrx * mtrx
- it transpose mtrx
- it premultiplies the current netwmat row by the newmat column.
- it returns the results of a matrix multiplication
- It squares each cell in mtrx
> newmat
[,1] [,2]
[1,] 9 25
[2,] 64 16
# The `%*%` operator gives matrix multiplication
> mtrx %*% mtrx
[,1] [,2]
[1,] 49 35
[2,] 56 56- connect()
- concat()
- contact()
- c()
- Rdefaults.site
- Renviron.site
- Rprofile.site
- Rstatus.site
- ncol(mdf) equals length(mdf).
- The number of rows must equals the number of columns.
- The legnth of any column in mdf may differ from any other column in mdf
- All columns must have the same data type.
Q38. A list can contain a list as an element. MyList has five columns, and the third column's item is a list of three items. How do you put all seven values in MyList into a single vector?
- vector(MyList, length = 7)
- coerce(MyList, nrows = 1)
- unlist(MyList)
- coerce(MyList, nrows = 7)
- ANOVAData, anovadata
- VisitPCA, VarX
- VisitPCA, varx
- Xvar, Yvar
StDf[, -1]- all but the first row and first column of StDf
- all but the final column of StDf
- all but the first column of StDf
- only the first column of StDf
- file.list()
- file.select()
- file.choose()
- file.open()
- Each is a type of data frame.
- Each is a type of atomic vector.
- Each is a type of complex vector.
- Each is a type of raw vector.
MyMat = matrix(c(7, 9, 8, 6, 10, 12),nrow=2,ncol=3, byrow = TRUE)- :
[ ,3]
[1, ] 8
[2, ] 12
- :
[1] 8 12
- :
[1] 10 12
- :
[ ,3]
[1, ] 10
[2, ] 12
- the probability of making a Type I error
- the probability of not making a Type II error
- the probability of making a Type II error
- the probability of not making a Type I error
result <- lm(outcome ~ covariate + factor + covariate:factor, data = testcoef)- It forces the intercepts of the individual regressions to zero.
- It calls for the effect of the covariate within each level of the factor.
- It calls for the effect of each variable from covariate to factor in testcoef.
- It forces the covariate to enter the equation before the factor levels.
# Example call to demonstrate. `Species` is a Factor. Petal.Length, Petal.Width are numeric.
# see `help(formula)` for more details on the formula specification. `:` is "effect modification" or "interaction"
> summary(lm(Petal.Length ~ Petal.Width + Species + Petal.Width:Species, data = iris))
...
Petal.Width:Speciesversicolor 1.3228 0.5552 2.382 0.0185 *
Petal.Width:Speciesvirginica 0.1008 0.5248 0.192 0.8480
...- integers and real values
- integers, real, and raw values
- real values only
- integers, real, and logical values
- property
- integer
- number
- variant
-
Rmat[upper.tri(Rmat)] -
upper.triangular(Rmat) -
upper.tri(Rmat) -
upper.diag(Rmat)
Q49. x is a vector of type integer, as shown on line 1 below. What is the type of the result returned by the statement > median(x)?
x <- c(12L, 6L, 10L, 8L, 15L, 14L, 19L, 18L, 23L, 59L)
- numeric
- integer
- single
- double
a <- list("10", TRUE, 5.6)
- is.list(a[1])
- is.numeric(a[1])
- is.logical(a[1])
- is.character(a[1])
Q51. How do you obtain the row numbers in a data frame named pizza for which the value of pizza$delivery_min is greater than or equal to 30?
- :
late_delivery <- pizza$delivery_min >= 30
index_late <- index(late_delivery)
index_late
- :
late_delivery <- pizza$delivery_min >= 30
rownum_late <- rownum(late_delivery)
rownum_late
- :
late_delivery <- pizza$delivery_min >= 30
which_late <- which(late_delivery)
which_late
- :
late_delivery <- pizza$delivery_min >= 30
late <- piza$late_delivery
pizza$late
indat <- c("Ash Rd","Ash Cir","Ash St")
- grepl("[Rd|Ave|Dr|St]", indat)
- grepl("Rd|Ave|Dr|St", indat)
- grepl("Rd,Ave,Dr,St", indat)
- grepl("[Rd],[Ave],[Dr],[St]", indat)
- fish[4, ]
- fish( ,4)
- fish(4, )
- fish{4, }
a <- c(1.2, 2, 3.5, 4)
b <- c(1.2, 2.2, 3.5, 4)
csum <-sum(a == b)
- 8
- 3
- 0.2
- 21.6
a <- list("10", TRUE, 5.6)
- is.list(a[1])
- is.numeric(a[1])
- is.logical(a[1])
- is.character(a[1])