-
Notifications
You must be signed in to change notification settings - Fork 2
/
README.Rmd
201 lines (166 loc) · 6.26 KB
/
README.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
---
output: rmarkdown::github_document
---
<!-- README.md is generated from README.Rmd. Please edit that file -->
```{r echo=FALSE, message=FALSE}
knitr::opts_chunk$set(message=FALSE, comment="#>")
devtools::load_all(".")
```
# shine
[]()
[](https://www.tidyverse.org/lifecycle/#maturing)
[](https://github.com/montilab/shine/commits/master)
**S**tructure Learning for **Hi**erarchical **Ne**tworks
*A package to aid in structure learning for hierarchical biological regulatory networks*
## Documentation
Please visit https://montilab.github.io/shine/ for comprehensive documentation.
## Requirements
We suggest R 3.6.0 but R (>= 3.5.0) is required to install directly from Github. For workflows, you will need Python (>= 2.7.0) and dependencies for [Nextflow](https://www.nextflow.io/). Nextflow can be used on any POSIX compatible system (Linux, OS X, etc) and requires BASH and Java 8 (or higher) to be installed. Alternatively, check out usage with Docker.
## Installation
Install the development version of the package from Github.
```{r, eval=FALSE}
devtools::install_github("montilab/shine")
```
```{r, eval=FALSE}
library(shine)
```
## Quick Example
```{r}
data(toy)
```
```
ABC
/ \
AB \
/ \ \
A B C
```
```{r}
dim(toy)
table(toy$subtype)
```
### Variable Selection
```{r}
# Filter out non-varying genes
genes.filtered <- keep.var(toy, column="subtype", subtypes=c("A", "B", "C"))
# Select top genes by median absolute deviation
genes.selected <- rank.var(toy, column="subtype", subtypes=c("A", "B", "C"), genes=genes.filtered, limit=75)
# Subset toy dataset
eset <- toy[genes.selected,]
```
### Structure Constraints
```{r, results='hide'}
# Detect modules
wgcna <- mods.detect(eset, min.size=5, cor.fn="cor", do.plot=FALSE)
```
```{r}
# Module membership
mods.plot(wgcna$dat, wgcna$mods, wgcna$colors, ncol=3, size=2.5)
```
```{r}
# Module extension
mods.extended <- fuzzy.mods(wgcna$dat, wgcna$mods, p=0.75)
mods.extended$grey <- NULL
mods <- sort(sapply(wgcna$mods, length), decreasing=TRUE)
print(mods)
sapply(mods.extended, length)[names(mods)]
```
### Network Estimation
```bash
$ curl -s https://get.nextflow.io | bash
```
--
*Hint*
Once downloaded make the `nextflow` file accessible by your $PATH variable so you do not have to specify the full path to nextflow each time. e.g. `nextflow run` rather than `path/to/nextflow run`
--
**Clone**
*See full documentation for [shine-nf](https://github.com/montilab/shine-nf).*
```bash
$ git clone https://github.com/montilab/shine-nf
```
**Docker**
```bash
$ docker pull montilab/shine:latest
```
**Hierarchy**
[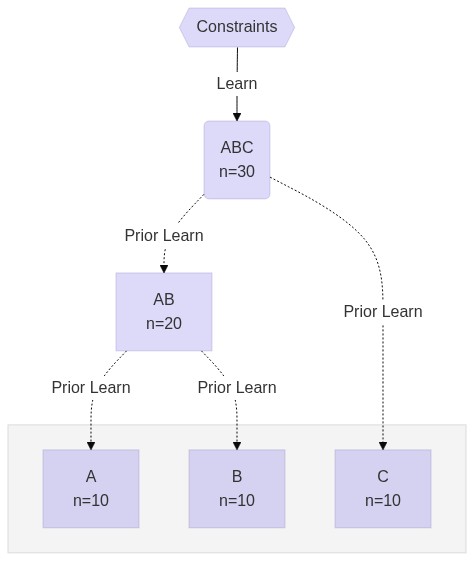](https://mermaid-js.github.io/mermaid-live-editor/#/edit/eyJjb2RlIjoiJSV7aW5pdDogeyd0aGVtZSc6ICdiYXNlJywgJ3NlY3VyaXR5TGV2ZWwnOiAnbG9vc2UnLCAndGhlbWVWYXJpYWJsZXMnOiB7ICdwcmltYXJ5Q29sb3InOiAnIzc4NjhlNjQwJywgJ2VkZ2VMYWJlbEJhY2tncm91bmQnOicjZmZmZmZmJywgJ3RlcnRpYXJ5Q29sb3InOiAnI2Y0ZjRmNCd9fX0lJVxuXG5ncmFwaCBURFxuICAgIFNDe3tDb25zdHJhaW50c319IC0tPiB8TGVhcm58IEFCQyhcIkFCQyA8YnIvPiBuPTMwXCIpXG4gICAgQUJDIC0uLT4gfFByaW9yIExlYXJufCBBQltcIkFCIDxici8-IG49MjBcIl1cbiAgICBBQiAtLi0-IHxQcmlvciBMZWFybnwgQVtcIkEgPGJyLz4gbj0xMFwiXVxuICAgIEFCIC0uLT4gfFByaW9yIExlYXJufCBCW1wiQiA8YnIvPiBuPTEwXCJdXG4gICAgQUJDIC0uLT4gfFByaW9yIExlYXJufCBDW1wiQyA8YnIvPiBuPTEwXCJdXG4gIFxuICAgIHN1YmdyYXBoIFwiIFwiXG4gICAgQVxuICAgIEJcbiAgICBDXG4gICAgZW5kXG4iLCJtZXJtYWlkIjp7InRoZW1lIjoiZGVmYXVsdCJ9LCJ1cGRhdGVFZGl0b3IiOmZhbHNlfQ)
**Define Workflow**
```sh
#!/usr/bin/env nextflow
# wf.nf
workflow ABC {
main:
eset = "data/esets/ABC.rds"
modules = "data/modules.rds"
SPLIT( eset, modules )
LEARN( SPLIT.out.flatten() )
RECONSTRUCT( eset, LEARN.out[0].collect() )
emit:
LEARN.out[0]
}
workflow AB {
take:
prior
main:
eset = "data/esets/AB.rds"
LEARN_PRIOR( eset, prior )
RECONSTRUCT( eset, LEARN_PRIOR.out[0].collect() )
emit:
LEARN_PRIOR.out[0]
}
workflow A {
take:
prior
main:
eset = "data/esets/A.rds"
LEARN_PRIOR( eset, prior )
RECONSTRUCT( eset, LEARN_PRIOR.out[0].collect() )
}
workflow B {
take:
prior
main:
eset = "data/esets/B.rds"
LEARN_PRIOR( eset, prior )
RECONSTRUCT( eset, LEARN_PRIOR.out[0].collect() )
}
workflow C {
take:
prior
main:
eset = "data/esets/C.rds"
LEARN_PRIOR( eset, prior )
RECONSTRUCT( eset, LEARN_PRIOR.out[0].collect() )
}
workflow {
ABC()
AB(ABC.out)
A(AB.out)
B(AB.out)
C(ABC.out)
}
```
**Run**
```bash
$ nextflow run wf.nf -with-docker montilab/shine
```
```
N E X T F L O W ~ version 20.07.1
Launching `wf.nf` [happy_koch] - revision: c2526aec9e
executor > local (36)
[f4/c715fd] process > ABC:SPLIT [100%] 1 of 1 ✔
[1b/37cc11] process > ABC:LEARN (6) [100%] 6 of 6 ✔
[99/23dcba] process > ABC:RECONSTRUCT [100%] 1 of 1 ✔
[1b/4e3537] process > AB:LEARN_PRIOR (6) [100%] 6 of 6 ✔
[2a/42f4df] process > AB:RECONSTRUCT [100%] 1 of 1 ✔
[da/17bb2f] process > A:LEARN_PRIOR (6) [100%] 6 of 6 ✔
[59/d209b2] process > A:RECONSTRUCT [100%] 1 of 1 ✔
[26/65e7a0] process > B:LEARN_PRIOR (6) [100%] 6 of 6 ✔
[7e/b65391] process > B:RECONSTRUCT [100%] 1 of 1 ✔
[58/721e46] process > C:LEARN_PRIOR (6) [100%] 6 of 6 ✔
[99/517354] process > C:RECONSTRUCT [100%] 1 of 1 ✔
Completed at: 21-Nov-2020 16:00:29
Duration : 2m 16s
CPU hours : 0.1
Succeeded : 36
```