Python Data Analysis Library
A fully-featured code library for manipulating data arranged in tables (i.e. matricies or data frames). It's arguably the most popular Python library used for data engineering. For those of you who have used the popular statistical language, R, Pandas brings many of that languages capabilities to python.
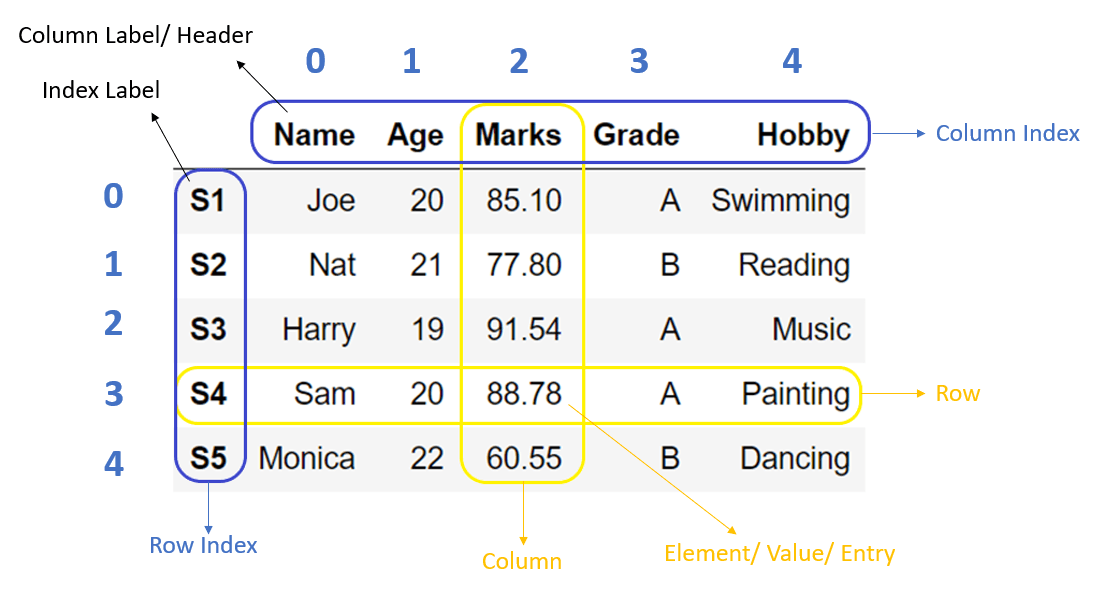
So far we've discussed how you build your own multidimensional objects like lists of lists and dictionaries of dictionaries from raw data. However, bioinformatics modules (and many others) will often return results in the form of Pandas data frame or a matrix. Further manipulation of these results (e.g. filtering, statistical analysis, data reorganization) will require some knowledge of Pandas operations.
-
Open a local file using Pandas, usually a comma-separated values (CSV) file, but could also be a tab-delimited text file (TSV), Excel, json, etc
-
Read a remote file on a website through a URL or read data from a remote database.
A matrix is an data structure where numbers are arranged into rows and columns. They will typically contain floats or integers, but not both. Matrices are used when you need to perform mathematical operations between datasets that contain multiple dimensions (i.e. measurements for two or more variables that change at the same time).
A data frame is a table-like data structure and can contain different data types (strings, floats, integers, etc.) in different columns. This is the type of data structure you're used seeing in Excel. Each column should only contain one data type.
Operations in Pandas, like R, work most efficiently when vectorized
You can think of a vector (also referred to as an array) as a type of list that contains a single data type and optimized for parallel computing. For matricies and data frames in Pandas (also NumPy), vectors are rows and columns.
Rather that looping through individual values (scalars), we apply operations to vectors (rows/columns). That is, the vector is treated as a single object. This topic can get a bit complicated, but it is worth doing your homework if you frequently work with these data types. Here's a few articles to get you started:
- A beginners guide to optimizing pandas code for speed.
- Why is vectorization faster in general than loops?
- Python Lists vs. Numpy Arrays, what's the difference?
Vectorization with Pandas series is ~390x faster than crude looping
Each function (method) in Pandas has many options and might not work the way you expect. It's definitely worth reading the documentation. Functions and options are sometimes updated, so even if you are already familiar with a function, it's a good idea to have a quick look.
Documentation is here https://pandas.pydata.org/docs/
Read getting started first https://pandas.pydata.org/docs/getting_started/index.html#getting-started
(what's possible: data types, summary stats, plots, table layouts, merging)
Pandas user guide (how it works, details on how Pandas thinks about data types) https://pandas.pydata.org/docs/user_guide/index.html#user-guide
Specific information about all the methods and classes https://pandas.pydata.org/docs/reference/index.html#api
But you'll probably want to start with a google search like pd load dataframeor pandas read excel skip rows
import pandas as pd
seq_info_input=pd.read_csv('sequencing_input.csv')
print(seq_info_input)
file_name library_id flowcell
s_1_1_CGATGT.fastq.gz L23058 H5K73BCX2
s_1_1_TGACCA.fastq.gz L23059 H5K73BCX2
s_1_1_ACAGTG.fastq.gz L23060 H5K73BCX2
#Read in a second dataframe for information transfer
sample_names=pd.read_csv("sample_names.csv")
library_id sample_name
L23058 E1
L23059 E2
L23060 E3
type(sample_names)
# prints <class 'pandas.core.frame.DataFrame'>
Note: We can read/write data in many other formats like tab delimited text .tsv and excel spreadsheets .xlsx. Please refer to this document for a full description of Pandas I/O tools.
You can build a dataframe from a dictionary file with df.DataFrame.from_dict(). By default the keys of the dictionary will become column names.
birthdays_dict={
'lab mate':['Carol', 'Vincent', 'Jin'],
'birthdate':['Sep 30', 'May 15', 'Feb 25']
}
print(birthdays_dict)
{'lab mate': ['Carol', 'Vincent', 'Jin'], 'birthdate': ['Sep 30', 'May 15', 'Feb 25']}
birthdays_df=pd.DataFrame.from_dict(birthdays_dict)
print(birthdays_df)
lab mate birthdate
0 Carol Sep 30
1 Vincent May 15
2 Jin Feb 25
Dataframes can be merged using identifiers that are present in both dataframes.
seq_info_input=pd.read_csv('sequencing_input.csv')
print(seq_info_input)
file_name library_id
s_1_1_CGATGT.fastq.gz L23058
s_1_1_TGACCA.fastq.gz L23059
s_1_1_ACAGTG.fastq.gz L23060
.. ..
s_1_1_ACAGTG.fastq.gz L23078
#Read in a second dataframe for information transfer
sample_names=pd.read_csv("sample_names.csv")
library_id sample_name body_length
L23058 E1 7.5
L23059 E2 7.7
L23060 E3 8.9
.. .. ..
L23878 E10 7.4
#Merge the two dataframes based on 'library_id' values
seq_info_input.merge(sample_names, on='library_id', how='inner')
file_name library_id sample_name body_length
s_1_1_CGATGT.fastq.gz L23058 E1 7.5
s_1_1_TGACCA.fastq.gz L23059 E2 7.7
s_1_1_TGACGG.fastq.gz L23060 E3 8.9
.. ... ..
s_1_1_ACAGTG.fastq.gz L23078 E10 7.4
If identifiers appear in one dataframe but not the other, you can select the acceptor and donor dataframes when merging.
"Slicing" refers to subsetting, or extracting rows and columns from a data frame.
Here's the general syntax to identify dataframe positions in [ rows , columns ] where rows and columns can be either labels or indices.
loc allows us to subset data by row or column label. For example, if I would like to pull out the column 'sample_name', I would use the following command:
print(seq_info_input)
file_name library_id sample_name body_length
s_1_1_CGATGT.fastq.gz L23058 E1 7.5
s_1_1_TGACCA.fastq.gz L23059 E2 7.7
s_1_1_TGACGG.fastq.gz L23060 E3 8.9
.. ... ..
s_1_1_ACAGTG.fastq.gz L23078 E10 7.4
[10 rows x 4 columns]
print(seq_info_input.loc[:,"sample_name"])
sample_name
0 E1
1 E2
2 E3
.. ..
10 E10
[10 rows x 1 columns]
iloc allows us to subset rows and colums by index number. This is useful if we want to subset multiple rows or columns without typing index names.
print(seq_info_input.iloc[:3])
body_length
7.5
7.7
8.9
..
7.4
[10 rows, 1 column]
print(seq_info_input.iloc[2,:])
Name: file_name, Length: 336, dtype: object
file_name s_1_1_TGACGG.fastq.gz
library_id L23059
sample_name E3
Name: 3, dtype: object
Note [[ ]] allows us to mention a list of columns.
# Return columns 0, 1, 3, 5, and 7
seq_info_input.iloc[:,[0,1,3,5,7]]
# Return rows 1 through 5 and columns 0, 1, 3, 5, and 7
seq_info_input.iloc[:5,[0,1,3,5,7]]
Here we'll take look at ordering our data by a particular column value, or multiple column values.
# Set ascending=True to reverse the order
seq_info_input.sort_values('sample_name', ascending=False)
# Sort by multiple columns in different directions
seq_info_input.sort_values(by=['sample_name', 'body_length'], ascending=[True, False])
Understanding how to subset your data using conditional operations is very, very useful. You'll often encounter situations where you want to filter your data on a certain set of parameters to reduce it to a more "meaningful" state.
# Subsetting on a single condition
seq_info_input.loc[(seq_info_input['body_length'] < 8 )]
In the example below we chain boolean operators together to achieve results that satisfy multiple conditions. You can make these statments complex as you'd like.
Note: Pandas uses the bitwise logical operators (see earlier lecture). A pipe symbol | represents or, and an ampersand symbol & represents and. The backslashes in code simply allow us to break up our statement at arbitrary points for readbility.
# Subsetting on multiple conditions.
seq_info_input[
(seq_info_input['body_length'] < 7) | \
(seq_info_input['sample_name'] == 'E3')]
What's actually going on here? The rows in the data frame are actually subsetted on a vector of True/False statements. That is, for every row for which the condition evaluates to True will be returned.
Subsetting might be the way you parse your RNA-seq results to a list of genes within a specific range of p-values and log fold changes, e.g., all p-values < 1e-15 and log fold changes > 1.2.
Lets look at a couple examples where we apply calculations to our data frame. First lets calculate some summary statistics. This can be a useful when viewing our results for the first time to get a handle on how our data is distributed.
# For this numerical data
data_df
A B C D E
0 45 38 10 60 76
1 37 31 15 99 98
2 42 26 17 23 78
3 50 90 100 56 90
# Returning summary statistics for all columns
data_df.describe()
A B C D E
count 4.000000 4.00000 4.000000 4.000000 4.000000
mean 43.500000 46.25000 35.500000 59.500000 85.500000
std 5.446712 29.57899 43.100657 31.118055 10.376255
min 37.000000 26.00000 10.000000 23.000000 76.000000
25% 40.750000 29.75000 13.750000 47.750000 77.500000
50% 43.500000 34.50000 16.000000 58.000000 84.000000
75% 46.250000 51.00000 37.750000 69.750000 92.000000
max 50.000000 90.00000 100.000000 99.000000 98.000000
# Returning summary statistics for a single column
data_df['A'].describe() # if you are working on a whole column
# Simply add the .corr() method to your dataframe subset
data_df.loc[:,['A','B']].corr()
A B
A 1.000000 0.830705
B 0.830705 1.000000
That summarizes our introduction to Pandas. As you can see, Pandas greatly simplifies the process of exploring and making calculations in data frames and matricies. Check out the link below for the official documentation.
Here's a few lines from a data file (fuelEfficiency.tsv)
| Mfr Name | Carline | Eng Displ | Cylinders | Transmission | CityMPG | HwyMPG | CombMPG | # Gears |
|---|---|---|---|---|---|---|---|---|
| aston martin | Vantage V8 | 4 | 8 | Auto(S8) | 18 | 25 | 21 | 8 |
| Volkswagen Group of | Chiron | 8 | 16 | Auto(AM-S7) | 9 | 14 | 11 | 7 |
| General Motors | CORVETTE | 6.2 | 8 | Auto(S8) | 12 | 20 | 15 | 8 |
| General Motors | CORVETTE | 6.2 | 8 | Auto(S8) | 15 | 25 | 18 | 8 |
| General Motors | CORVETTE | 6.2 | 8 | Auto(S8) | 14 | 23 | 17 | 8 |
It has 718 lines of data and a header line.
Let's examine the data in the file: data types, inconsistencies. What about trends in values?
We can use the python interpreter
>>> import pandas as pd
>>> import matplotlib.pyplot as plt
>>> df = pd.read_csv('fuelEfficiency.tsv', sep='\t')
>>> df
Mfr Name Carline Eng Displ Cylinders Transmission CityMPG HwyMPG CombMPG # Gears
0 aston martin Vantage V8 4.0 8 Auto(S8) 18 25 21 8
1 Volkswagen Group of Chiron 8.0 16 Auto(AM-S7) 9 14 11 7
2 General Motors CORVETTE 6.2 8 Auto(S8) 12 20 15 8
3 General Motors CORVETTE 6.2 8 Auto(S8) 15 25 18 8
4 General Motors CORVETTE 6.2 8 Auto(S8) 14 23 17 8
.. ... ... ... ... ... ... ... ... ...
713 Toyota 4RUNNER 4WD 4.0 6 Auto(S5) 17 20 18 5
714 Toyota LAND CRUISER WAGON 4WD 5.7 8 Auto(S8) 13 18 15 8
715 Toyota SEQUOIA 4WD 5.7 8 Auto(S6) 13 17 14 6
716 Volvo XC90 AWD 2.0 4 Auto(S8) 19 26 22 8
717 Volvo XC90 AWD 2.0 4 Auto(S8) 20 27 23 8
To get very useful summary statistics, use df.describe()
>>> df.describe()
Eng Displ Cylinders CityMPG HwyMPG CombMPG # Gears
count 718.000000 718.000000 718.000000 718.000000 718.000000 718.000000
mean 3.092061 5.493036 20.442897 27.760446 23.139276 7.147632
std 1.344572 1.752251 5.298504 5.607924 5.368443 1.507929
min 1.400000 3.000000 9.000000 14.000000 11.000000 1.000000
25% 2.000000 4.000000 17.000000 24.000000 19.000000 6.000000
50% 3.000000 6.000000 20.000000 27.000000 23.000000 7.000000
75% 3.600000 6.000000 23.000000 31.000000 26.000000 8.000000
max 8.000000 16.000000 57.000000 59.000000 58.000000 10.000000
We can make simple plots with pandas and matplotlib
We can show the distribution and range of a single set of data in a histogram.
Here are the bare bones, you can add/customize parameters yourself.
#!/usr/bin/env python3
import pandas as pd
import matplotlib.pyplot as plt # we will use this plotting library
# read data into a pandas dataframe
df = pd.read_csv('fuelEfficiency.tsv', sep='\t')
fig,ax=plt.subplots(figsize=(6,6))
ax.hist(df['Cylinders'], bins=10)
# set title, axis labels
ax.set_xlabel('Number of cylinders')
ax.set_ylabel('Count')
ax.set_title('Vehicle fuel efficiency data')
#write histogram to a PNG graphics file
png_file = 'cylinders.hist.png'
fig.savefig(png_file)
print(f'Wrote {png_file}')
Here's the plot
These are great for looking at a relationship between two measurements or values.
#!/usr/bin/env python3
import pandas as pd
import matplotlib.pyplot as plt # we will use this plotting library
fig,ax=plt.subplots(figsize=(6,6))
# set transparency with alpha and data point size with s
ax.scatter(df['Cylinders'],df['HwyMPG'], alpha = 0.5, s=2)
# set title, axis labels
ax.set_xlabel('Number of cylinders')
ax.set_ylabel('Highway Miles per Gallon')
ax.set_title('Vehicle fuel efficiency data')
#write histogram to a PNG graphics file
png_file = 'cylindersVShwyMPG.scatter.png'
fig.savefig(png_file)
print(f'Wrote {png_file}')
Here's the scatter plot. You can clearly see a relationship as well as variability in the data.
What's another good way to investigate a relationship between two sets of data?
That summarizes our introduction to Pandas. As you can see, Pandas greatly simplifies the process of exploring and making calculations in data frames and matricies. Check out the link below for the offical documentation.