This is the official code repository for "Classification of Gleason Grading in Prostate Cancer Histopathology Images Using Deep Learning Techniques: YOLO, Vision Transformers, and Vision Mamba"
| Dataset | Task | precision | Sensitivity | Specificity | F1-score | Overall Accuracy | AUC | Model Weight |
|---|---|---|---|---|---|---|---|---|
| Gleason2019 | Multi-Class(4) | TBA | TBA | TBA | TBA | TBA | TBA | Coming soon! |
| SICAPv2 | Multi-Class(4) | TBA | TBA | TBA | TBA | TBA | TBA | Coming soon! |
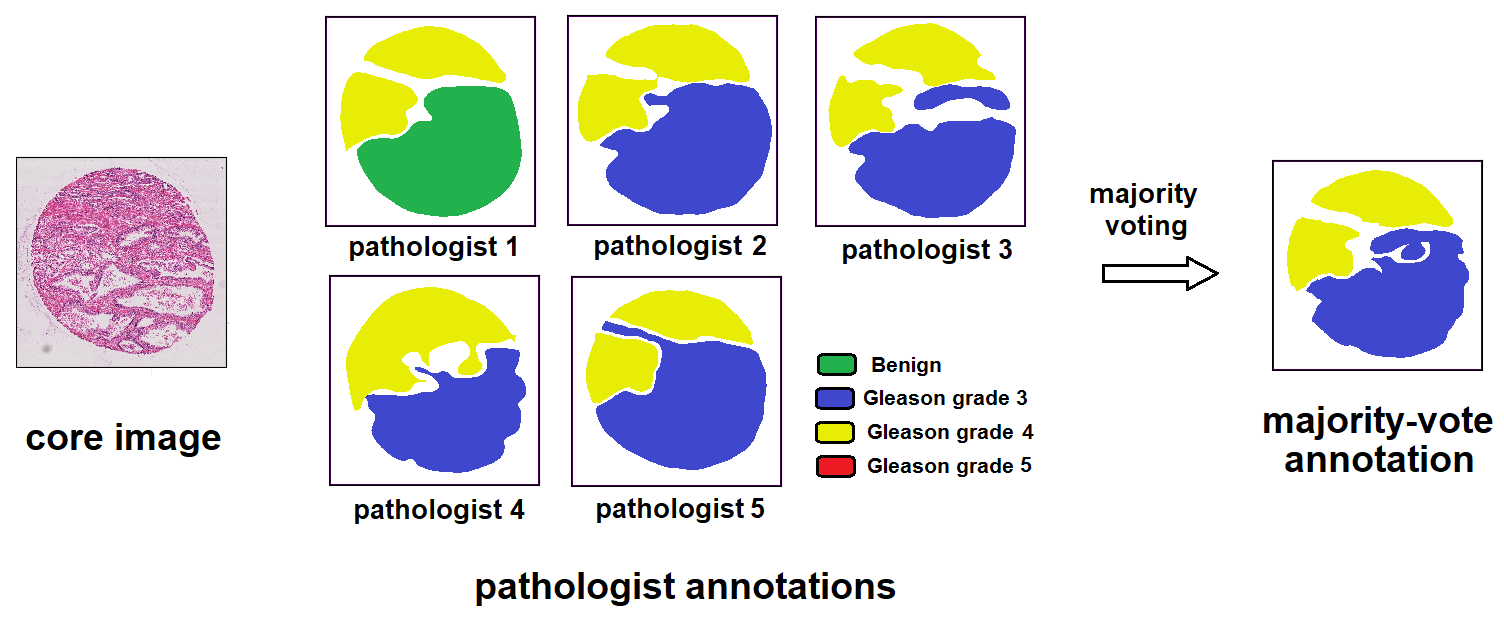
The Gleason 2019 dataset comprises TMA images with a resolution of 5120 x 5120 pixels, which are annotated by several experts. After calculating the final annotation map for each image by pixel-level majority voting, we opt for smaller patches inside original images to train our model. Specifically, we extract 512 x 512 patches with a stride of 256 in each image, and assign a single label for them based on pixel annotations at their central areas. If all of the pixels residing in this 250 x 250 area do not belong to the background and share an identical label, the patch label is set accordingly, otherwise the patch is discarded.Gleason2019 Dataset
The SicapV2 dataset offers whole slide histology images with both global Gleason scores and patch-level grades. Like the Gleason2019 dataset, the patches are 512 x 512 pixels with a 50% overlap. The authors offer a meticulous patient-based split of this data, which guaranteed the class balance between cross-validation subsets. .SICAPv2 Dataset
We thank the authors of medmamba for their open-source codes.