-
Notifications
You must be signed in to change notification settings - Fork 4
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Add functionality to improve explainability to tidymodels #11
Comments
|
Those packages would just need to make some changes to the code that makes predictions. I suspect that I don't think that we would make any packages in this area since the existing ones cover all of the ground. The best thing that you can do is to put in |
|
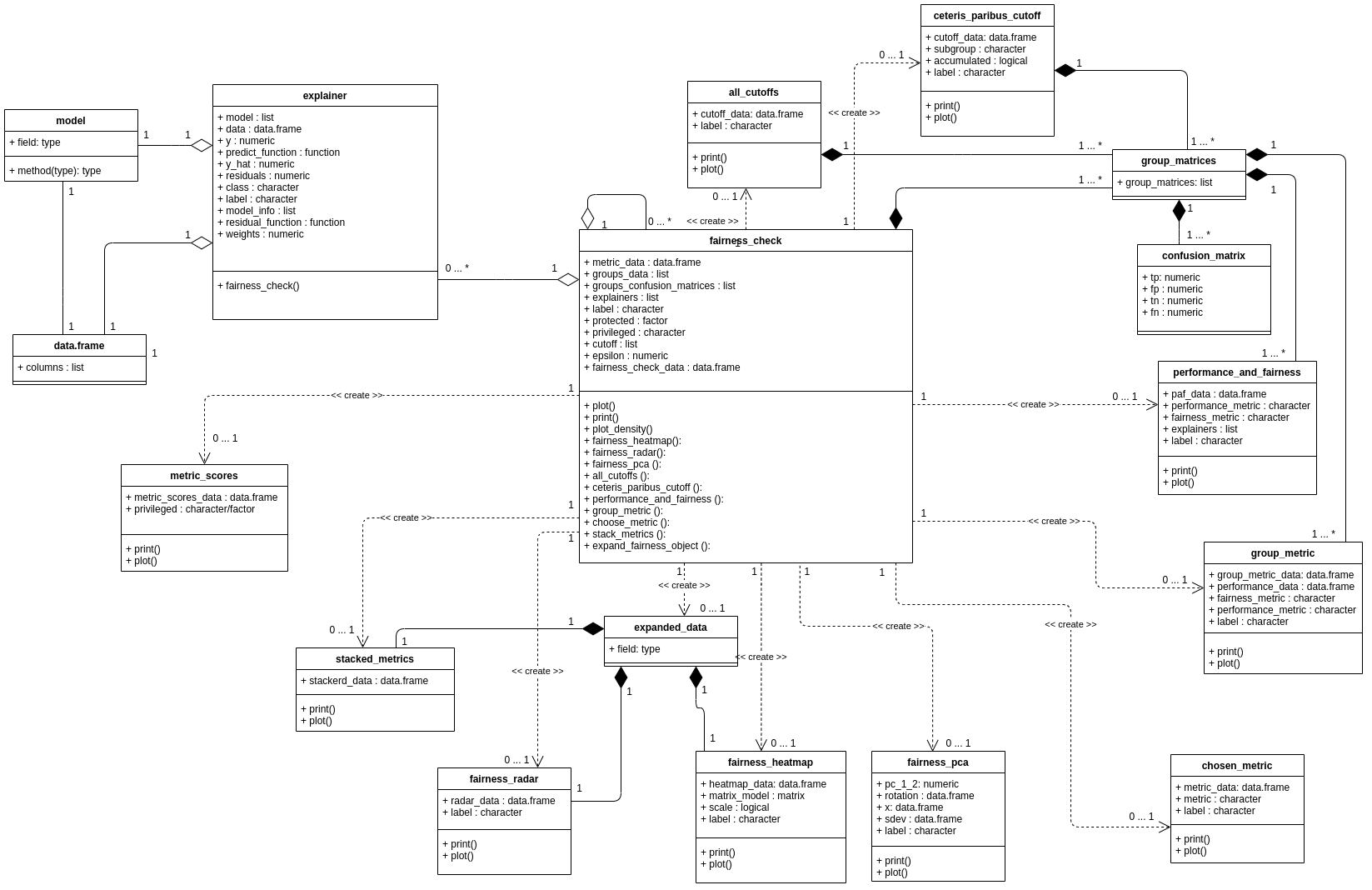
Thanks, The plan is to prepare a tutorial on how to use these packages together, something like this chapter for DALEX+mlr3 https://mlr3book.mlr-org.com/interpretability-dalex.html. @topepo It would be fantastic to have for tidymodels a documentation of the object interfaces. https://github.com/ModelOriented/fairmodels/blob/master/man/figures/class_diagram.png |
|
Is there a tool that generates the diagram from a codebase (or other source)? I don't know that we will generate this by hand. That's said, I can give you any details that you need regarding interfaces. |
|
Also, we would be very happy to include a |
|
Sounds great! A. the task that is being performed (classification, regression, something else), As far as data is concerned, I understand that there are at least two stages: raw and after processing with recipes. I'm guessing that the predict function understands data after processing with recipes. If the model does not contain the training data by itself, I would need some mechanism to provide the data in the form that the predict function understands. Maybe it is possible to draw from the model an object describing pre-processing steps from the receptions. In the scikit learn, the pipeline contains both variable transformations and the model, so there the predict/predict_proba function can operate on raw data. The model itself does not contain data, so we supply them independently in the raw form. Pipeline's predict function understands how to work on raw data. As far as UML diagrams are concerned, unfortunately, they are still made manually. We want to automate it somehow, but because S3 classes don't have strictly defined structures it's not easy. |
|
Here's some technical details for your questions... library(tidymodels)What is the task being performed?Our models contain a mode attribute that describes the type of problem. Currently, the modes are "unknown", "classification", and "regression". More modes will follow when we start integrating survival analysis models. Before being assigned, the mode for a model specification is "unknown". A specification cannot be fit without an unknown mode. Here's an example: knn_mod <-
nearest_neighbor(neighbors = 3) %>%
set_engine("kknn") %>%
set_mode("regression")For knn_mod$modeFor a workflow, you pull the model specification and get the mode: knn_workflow <-
workflow() %>%
add_model(knn_mod) %>%
add_formula(mpg ~ .)
knn_workflow %>%
workflows::pull_workflow_spec() %>%
pluck("mode")Once the model is fit, the process is similar since the specification is a sub-element of the fit: # parsnip:
knn_mod %>%
fit(mpg ~ ., data = mtcars) %>%
pluck("spec") %>%
pluck("mode")# workflows:
knn_workflow %>%
fit(data = mtcars) %>%
workflows::pull_workflow_fit() %>%
pluck("spec") %>%
pluck("mode")Interface to the predict functionThere is one consistent interface to the predict methods: The main rules that makes it different than base R predict functions are:
Again, the prediction interface is the same for For example: knn_mod %>%
fit(mpg ~ ., data = mtcars) %>%
predict(mtcars %>% head())knn_workflow %>%
fit(data = mtcars)%>%
predict(mtcars %>% head())A classification example with data("two_class_dat", package = "modeldata")
levels(two_class_dat$Class)glm_fit <-
logistic_reg() %>%
set_engine("stan") %>%
# Mode is automatically set for models with only one possible value
fit(Class ~ ., data = two_class_dat)
predict(glm_fit, two_class_dat %>% head()) # 'class' is the default typepredict(glm_fit, two_class_dat %>% head(), type = "prob")predict(glm_fit, two_class_dat %>% head(), type = "conf_int")predict(glm_fit, two_class_dat %>% head(), type = "pred_int")Again, workflow objects have the same syntax. This probably won't matter to your applications, but some model predictions can produce nested data frame columns. For example, if someone is doing quantile regression and wants 7 specific quantiles, the tibble produced by Data that were used for training for the modelWe generally eschew saving the training set with the model objects (along with pre-computed things like residuals and fitted values). For large data sets, this is problematic and all of these values can be re-produced on demand. This is pretty consistent with the S language belief in laziness. The model object itself might save these, but that is model dependent. One other reason is that, especially for recipes, the data used to fit the model are not the original data. If we do any pre-processing or feature engineering, we strongly suggest keeping that separate from the model object.
Yes, this. This separation of data manipulation and model fitting might impact you because someone would give you a That's why most of our focus is on workflows; these bundle together the pre-process/feature engineering/data processing with the model. For example, if we want to do PCA prior to the model: pca_rec <-
recipe(Class ~ ., data = two_class_dat) %>%
step_normalize(all_predictors()) %>%
step_pca(all_predictors(), num_comp = 2)
glm_mod <-
glm_fit <-
logistic_reg() %>%
set_engine("stan")
glm_pca_wflow <-
workflow() %>%
add_model(glm_mod) %>%
add_recipe(pca_rec)
# does the PCA and normalization steps before modeling:
glm_pca_fit <-
glm_pca_wflow %>%
fit(data = two_class_dat)
glm_pca_fitThen predict(glm_pca_fit, two_class_dat %>% head(), type = "prob")I know that you have a page on using Unique name/identifier of the modelWe don't really have that. |
|
Thank you. We'll take a closer look at these options |
|
No problem. Let me know if you have more questions. |
|
Thank you so much for your replies! Great to hear that there are plans to make it easier to integrate DALEX with tidymodels. |
|
@Sponghop but it would be a great test if you could prepare a model with some advanced workflow for the Diabetes data. We would see if the solution for titanic can also be applied to the model you build |
|
I made an example that uses recipes that could work as a template for a In this example, the main predictor is a date column that gets engineered by a recipe into different features. Most everything in Anyway, give it a look and let me know what you think. |
|
@pbiecek this the link to the code we created for combining tidymodels with Dalex: https://github.com/Sponghop/dalex_tidymodels_example We would be glad to hear your opinion about it and if we can be of further assistance. |
|
@Sponghop @topepo Thank You for those extensive examples! I've read through them and many things about how that integration should work are clear now. It looks doable to add tidymodels support without much interference in source code. I Will let You know once I finish branch with it and allow You to test it. |
|
Thanks guys for all your examples. @maksymiuks has prepared Sponghop, examples with auto-tuned models are great. I still have to think whether it is possible to squeeze more in DALEX from data about the grid of tested models. Here are some examples of 0. Prepare data1. Explanations operate on original feature spacePrepare recipes and train models. Create an explainer Try local and global explanations 2. Explanations operate on transformed feature space, after recipePrepare data (bake) and model Create an explainer Try global and local explanations 3. Explain two or model models in a single plot (Rashomon perspective)Let's add Ranger model and explainer. And put both models in a plot. |
It would be great when methods for making models and predictions better explainable would be made available through tidymodels and could be added to a workflow, for example like the ones in the
DALEXandimlpackage. Are there any plans to do this?The text was updated successfully, but these errors were encountered: