forked from datajoint/datajoint-python
-
Notifications
You must be signed in to change notification settings - Fork 0
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Merge pull request #1 from A-Baji/horst-changelog
pull from upstream and update changelog
- Loading branch information
Showing
9 changed files
with
79 additions
and
26 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,7 @@ | ||
| # Citation | ||
|
|
||
| If your work uses the DataJoint API for Python, please cite the following manuscript and Research Resource Identifier (RRID): | ||
|
|
||
| - Yatsenko D, Reimer J, Ecker AS, Walker EY, Sinz F, Berens P, Hoenselaar A, Cotton RJ, Siapas AS, Tolias AS. DataJoint: managing big scientific data using MATLAB or Python. bioRxiv. 2015 Jan 1:031658. doi: https://doi.org/10.1101/031658 | ||
|
|
||
| - DataJoint API for Python - [RRID:SCR_014543](https://scicrunch.org/resolver/SCR_014543) - Version `Enter version here` |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,36 @@ | ||
| # Welcome to the DataJoint API for Python! | ||
|
|
||
| The DataJoint API for Python is a framework for scientific workflow management based on relational principles. DataJoint is built on the foundation of the relational data model and prescribes a consistent method for organizing, populating, computing, and querying data. | ||
|
|
||
| DataJoint was initially developed in 2009 by Dimitri Yatsenko in Andreas Tolias' Lab at Baylor College of Medicine for the distributed processing and management of large volumes of data streaming from regular experiments. Starting in 2011, DataJoint has been available as an open-source project adopted by other labs and improved through contributions from several developers. | ||
| Presently, the primary developer of DataJoint open-source software is the company [DataJoint](https://datajoint.com){:target="_blank"}. | ||
|
|
||
| ## Data Pipeline Example | ||
|
|
||
| 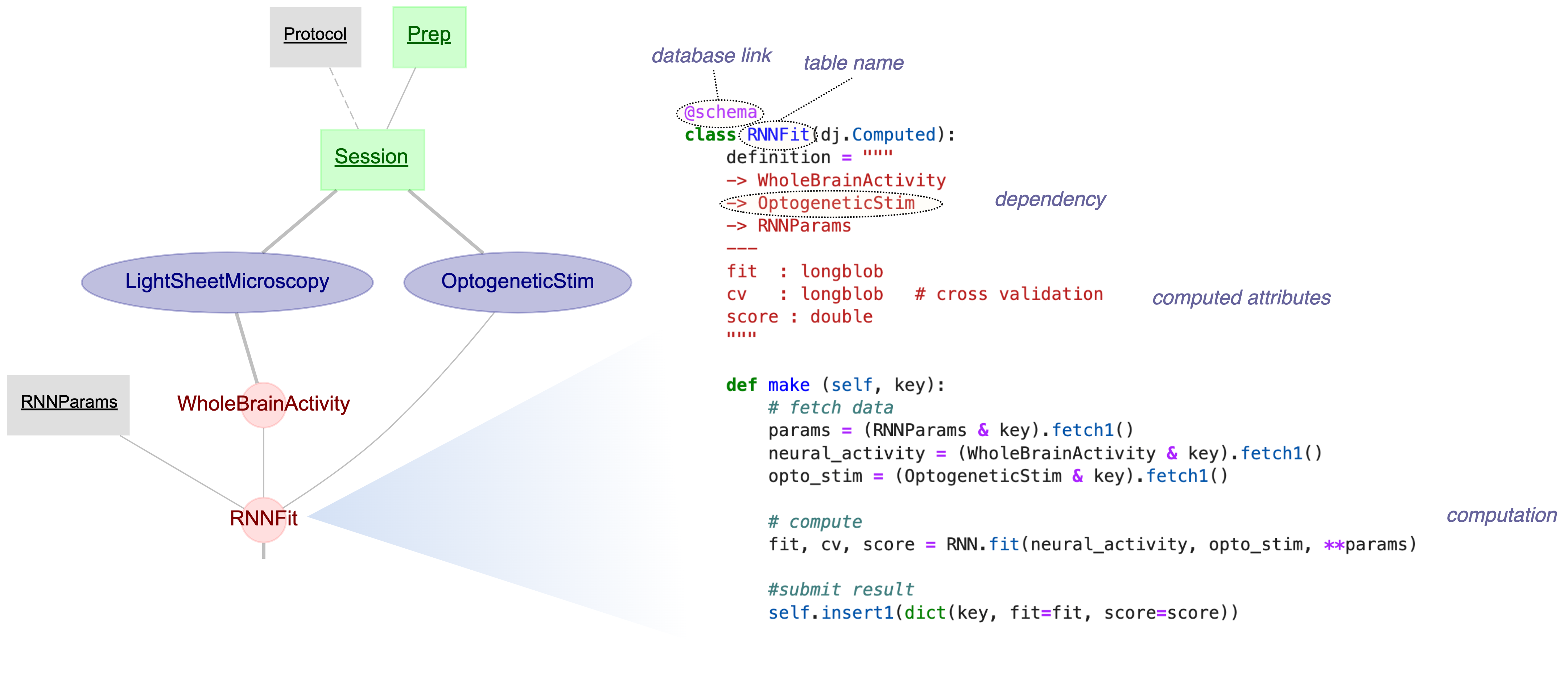 | ||
|
|
||
| [Yatsenko et al., bioRxiv 2021](https://doi.org/10.1101/2021.03.30.437358){:target="_blank"} | ||
|
|
||
| ## Getting Started | ||
|
|
||
| - Install from PyPI | ||
|
|
||
| ```bash | ||
| pip install datajoint | ||
| ``` | ||
|
|
||
| - [Detailed Getting Started Guide](./getting-started) | ||
|
|
||
| - [Interactive Tutorials](https://github.com/datajoint/datajoint-tutorials){:target="_blank"} on GitHub Codespaces | ||
|
|
||
| - [DataJoint Elements](../../elements/) - Catalog of example pipelines for neuroscience experiments | ||
|
|
||
| - Contribute | ||
| - [Development Environment](./develop) | ||
|
|
||
| - [Guidelines](../../community/contribute/) | ||
|
|
||
| - Legacy Resources (To be replaced by above) | ||
| - [Documentation](https://docs.datajoint.org) | ||
|
|
||
| - [Tutorials](https://tutorials.datajoint.org) |
Large diffs are not rendered by default.
Oops, something went wrong.
Loading
Sorry, something went wrong. Reload?
Sorry, we cannot display this file.
Sorry, this file is invalid so it cannot be displayed.