Where I share fabulous data science, biostatistics, informatics, and data science resources while teaching myself new ways to present information using Markdown!!
- OHDSI_Resources
- OMOP CDM Basic Data Dictionary
- Projects Best Suited for Observational Research and OHDSI Network Studies
- Analytic Use Cases and Examples
- [Data Management Tools and Resources(##Data-Management-Tools-and-resources)
- Incremental Loading
- The Collaboration Process
- Data Content Ontology
- Current CDM
- Commonly Used CDM Tables Overview
- ETL Basics
- OHDSI Analysis Tools
- Data Science Handbook
- Jupyter Notebooks and programming
- Recommended Trainings
- Python, SQL, and R Programming Resources
- Analysis with SQL
- Analysis with R
- Diversity, Equity, and Inclusion Resources
- [REDCAP training resources] (#redcap-training-resources)
For a sample interactive OMOP data dictionary, please click on the image below:
| Analytic use case | Type | Structure | Example |
|---|---|---|---|
| Clinical characterization | Disease Natural History | Amongst patients who are diagnosed with <insert your favorite disease>, what are the patient’s characteristics from their medical history? |
Amongst patients with rheumatoid arthritis, what are their demographics (age, gender), prior conditions, medications, and health service utilization behaviors? |
| Treatment utilization | Amongst patients who have <insert your favorite disease>, which treatments were patients exposed to amongst <list of treatments for disease> and in which sequence? |
Amongst patients with depression, which treatments were patients exposed to SSRI, SNRI, TCA, bupropion, esketamine and in which sequence? | |
| Outcome incidence | Amongst patients who are new users of <insert your favorite drug>, how many patients experienced <insert your favorite known adverse event from the drug profile> within <time horizon following exposure start>? |
Amongst patients who are new users of methylphenidate, how many patients experienced psychosis within 1 year of initiating treatment? | |
| Population-level effect estimation | Safety surveillance | Does exposure to <insert your favorite drug> increase the risk of experiencing <insert an adverse event> within <time horizon following exposure start>? |
Does exposure to ACE inhibitor increase the risk of experiencing Angioedema within 1 month after exposure start? |
| Comparative effectiveness | Does exposure to <insert your favorite drug> have a different risk of experiencing <insert any outcome (safety or benefit)> within <time horizon following exposure start>, relative to <insert your comparator treatment>? |
Does exposure to ACE inhibitor have a different risk of experiencing acute myocardial infarction while on treatment, relative to thiazide diuretic? | |
| Patient level prediction | Disease onset and progression | For a given patient who is diagnosed with <insert your favorite disease>, what is the probability that they will go on to have <another disease or related complication> within <time horizon from diagnosis>? |
For a given patient who is newly diagnosed with atrial fibrillation, what is the probability that they will go onto to have ischemic stroke in next 3 years? |
| Treatment response | For a given patient who is a new user of <insert your favorite chronically-used drug>, what is the probability that they will <insert desired effect> in <time window>? |
For a given patient with T2DM who start on metformin, what is the probability that they will maintain HbA1C <6.5% after 3 years? | |
| Treatment safety | For a given patient who is a new user of <insert your favorite drug>, what is the probability that they will experience <insert adverse event> within <time horizon following exposure>? |
For a given patient who is a new user of warfarin, what is the probability that they will have GI bleed in 1 year? |
This document provides a detailed overview of several essential clinical terminologies and coding systems used in healthcare. Each system has a specific role and is crucial for standardized communication in healthcare settings. The information includes development history, usage, and updates of these systems.
For more in-depth information, links to the respective official websites are provided.
- Development: Originally by the College of American Pathologists, now under SNOMED International.
- Adoption: Used in over 50 countries.
- Concepts: Over 340,000 active concepts in 19 hierarchies.
- Usage: Encodes clinical information including diseases, findings, and procedures.
- Updates: Biannual, with more frequent updates planned.
- More Information: SNOMED International
- Developer: Regenstrief Institute.
- Function: Identifiers for laboratory and clinical observations.
- Content: Over 90,000 terms.
- Collaboration: With SNOMED CT for coded content development.
- Updates: Biannual.
- More Information: LOINC
- Developer: National Library of Medicine (NLM).
- Function: Standard nomenclature for medications.
- Integration: Links to various drug vocabularies.
- Access: Requires UMLS user license for proprietary content.
- More Information: RxNorm - NLM
- Endorsement: World Health Organization (WHO).
- Versions: ICD-10 widely used with national extensions; ICD-11 adopted for future use.
- Purpose: Epidemiology, health management, clinical purposes.
- Updates: Annual, freely available.
- More Information: WHO ICD
- Developer: American Medical Association (AMA).
- Use: Encoding of medical services and procedures in the USA.
- Categories: Three categories of codes.
- Requirement: License from AMA for use.
- More Information: CPT - AMA
- Function: Bioinformatic resources for human diseases and phenotypes analysis.
- Components: Phenotype vocabulary, disease-phenotype annotations, algorithms.
- Applications: Genomic interpretation, gene-disease discovery, precision medicine.
- Content: Over 13,000 terms in 5 hierarchies.
- Availability: Freely available, multiple releases per year.
- More Information: Human Phenotype Ontology
- Initiation: By the US National Library of Medicine in 1986.
- Goal: To aid in the retrieval and integration of electronic biomedical information.
- Challenge Addressed: Different vocabularies expressing the same information differently.
- Availability: Free, but requires a license due to additional licensing requirements of some contents.
- More Information: UMLS - NLM
-
Process: Finding the closest match of a code from one ontology in another.
-
Matching: Exact equivalence is rare; approximate matching is common.
-
Challenges: Labor-intensive and requires understanding the maps' nature and limitations.
-
Alternative Approach: Mapping multiple ontologies to a central core terminology, as used by the OHDSI consortium.
-
More Information: BioPortal
graph LR
ICD9("ICD9") -->|Transformation to OMOP CDM| SNOMED("STANDARD<br>Vocabulary Concept Code<br>SNOMED")
ICD10("ICD10") -->|Transformation to OMOP CDM| SNOMED
| Domain | Source Vocabulary | Standard Vocabulary |
|---|---|---|
| Conditions | ICD9, ICD10 | SNOMED |
| Measurements | LOINC or institutional specific codes | LOINC |
| Drugs | NDC | RxNORM |
| Procedures | ICD9, ICD10, CPT | SNOMED |
-
ICD = International Classification of Diseases
-
SNOMED = Systematized Nomenclature of Medicine
-
LOINC = Logical Observation Identifiers Names and Codes
-
NDC = National Drug Code
-
CPT = Current Procedural Terminology
Incremental loading in the context of OHDSI refers to the process of adding new or updated data to an existing OHDSI database without the need to completely rebuild or refresh the entire dataset. This can be particularly useful for large datasets where full loads can be time-consuming and inefficient. The process involves extracting only the changes since the last load and then transforming and loading this delta of data into the existing OMOP Common Data Model (CDM) used by OHDSI tools.
For instance, in the development of an ETL (Extract, Transform, Load) process for the bulk and incremental load of German patient data into the OMOP CDM using FHIR as referenced by OHDSI, it suggests that the incremental loading is an essential part of keeping the database up-to-date in an efficient manner. OHDSI Symposium Showcase #44
This group alos described a Near Real-Time Incremental OMOP-CDM ETL System
This is also described by Dr. DuWayne Willett, CMIO of UTSW, at around minute 30 of this video:
...and in this OHDSI symposium presentation: 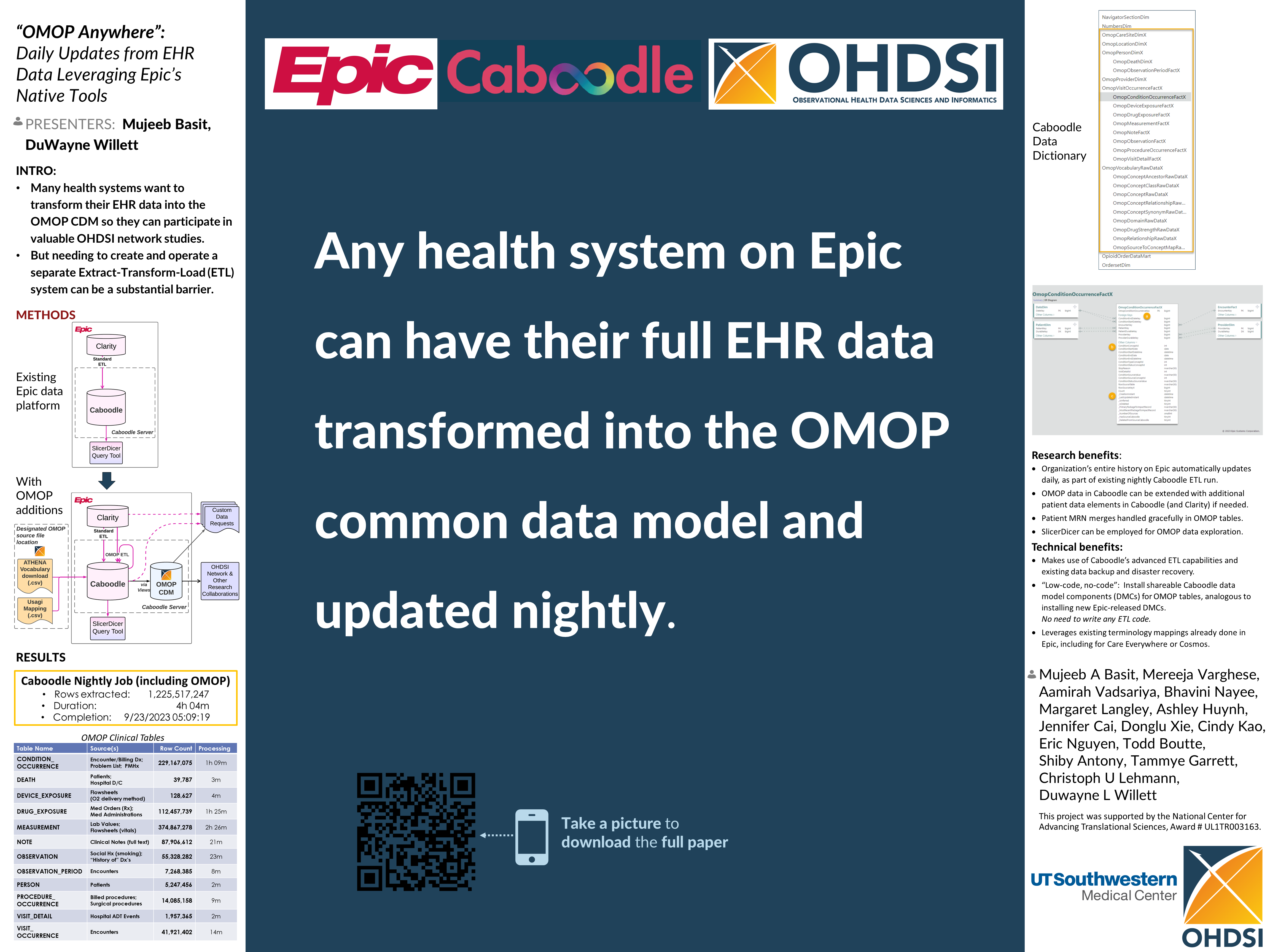
flowchart TD
A[Person A receives a research request]
B[Person/group B sets up a meeting]
C[Iterative biomedical query mediation process begins]
D[Project outline is created and signed off by all parties]
E[Voucher/payment/estimate is produced including resource and timeframe]
F[Department head reviews/signs off]
G[Work begins]
H[First draft is produced with a notebook outlining results]
I[Meeting is scheduled to review results of the notebook]
J[Necessary modifications are made and returned to the researcher]
K[End of Process]
A --> B
B --> C
C --> D
D --> E
E --> F
F --> G
G --> H
H --> I
I --> J
J -.->|If modifications needed| G
J -->|If complete| K
Source: OHDSI Common Data Model
Interactive (Select) OMOP Data Dictionary
The OMOP common data model (CDM) is a relational database made up of different tables that relate to each other by foreign keys (XXXX_ID values; e.g., PERSON_ID or PROVIDER_ID). The OMOP tables in your data export are as follows:
| Table | Description |
|---|---|
| Person | Contains basic demographic information describing a participant, including biological sex, birth date, race, and ethnicity. |
| Visit_occurrence | Captures encounters with healthcare providers or similar events. Contains the type of visit a person has (outpatient care, inpatient care, or long-term care), as well as the date and duration information. Rows in other tables can reference this table, for example, condition_occurrences related to a specific visit. |
| Condition_occurrence | Indicates the presence of a disease or medical condition stated as a diagnosis, a sign, or symptom, which is either observed by a provider or reported by the patient. |
| Drug_exposure | Captures records about the utilization of a medication. Drug exposures include prescription and over-the-counter medicines, vaccines, and large-molecule biologic therapies. Radiological devices ingested or applied locally do not count as drugs. Drug exposure is inferred from clinical events associated with orders, prescriptions written, pharmacy dispensing, procedural administrations, and other patient-reported information. |
| Measurement | Contains both orders and results of a systematic and standardized examination or testing of a participant or participant's sample, including laboratory tests, vital signs, quantitative findings from pathology reports, etc. |
| Procedure_occurrence | Contains records of activities or processes ordered by or carried out by a healthcare provider on the patient to have a diagnostic or therapeutic purpose. |
| Observation | Captures clinical facts about a person obtained in the context of an examination, questioning, or a procedure. Any data that cannot be represented by another domain, such as social and lifestyle facts, medical history, and family history, are recorded here. |
| Device_exposure | Captures information about a person's exposure to a foreign physical object or instrument which is used for diagnostic or therapeutic purposes. Devices include implantable objects, blood transfusions, medical equipment and supplies, other instruments used in medical procedures, and material used in clinical care. |
| Death | Contains the clinical events surrounding how and when a participant dies. |
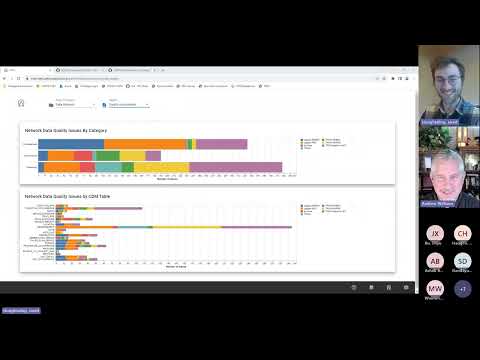
The Book of OHDSI - Chapter 15: Data Quality
https://www.ohdsi.org/wp-content/uploads/2019/09/OMOP-Common-Data-Model-Extract-Transform-Load.pdf https://ohdsi.github.io/TheBookOfOhdsi/ExtractTransformLoad.html
-
Dataset profiling and documentation
- Create data model documentation, sample data, data dictionaries, code lists, and other relevant information (23-Aug)
- Execute database profiling scan (WhiteRabbit) on source database
- Prepare mapping approach/documents based on scan reports from database profiling scan
-
Generation of the ETL Design
- Mapping workshop with all relevant parties to:
- Understand the source
- Define the scope of source data to be transformed
- Define acceptance criteria for OMOP output
- Output: draft mapping document
- Finalize mapping document:
- Integrate all notes/documentation from workshop
- Work through mappings and verify, update, fill in gaps
- Meetings/emails with data contact/technical contact (TC) as needed
- Mapping workshop with all relevant parties to:
-
Source Data Integrations and Semantic Mapping
- Source Code mapping:
- Identify which codes are already mapped to standard vocabulary
- Identify code types for codes that need to be mapped
- Translation of code description/phrases to English, if/as needed
- Create proposed code mappings
- Generate mappings for data coming out of flowsheets (together with consortium)
- Review/approval of code mappings, often done by medical experts affiliated with Data Owner (DO).
- Identify medical imaging available and define mappings to Imaging Extension
- Identify waveform data available and map using consortium-defined guidelines
- Use OHNLP to extract OMOP data from unstructured sources
- Source Code mapping:
-
Technical architecture design
- Continuous Integration, Continuous Deployment (CI/CD):
- Decide on ETL dev/deployment flow
- Put version control mechanisms in place
- OHDSI Ecosystem:
- Evaluate infrastructure needed
- Create infrastructure design documentation
- Continuous Integration, Continuous Deployment (CI/CD):
-
Technical ETL Development
- Implement ETL (Preferred Language/Structure?)
- Update ETL based on testing/QA/feedback (8, 9)
-
Setting up of Infrastructure
- Deploy core servers and associated services based on infrastructure design in (4)
-
Installation of the OHDSI tools
- Install and configure all software (database server, Achilles/DQD/Ares, Atlas/WebAPI, R Studio server, HADES, notebooks/tooling related to analytics, and any other software to suit a site’s specific needs).
-
ETL Testing and Validation
- ETL Execution:
- Test ETL using sample/development data (with limited external data access)
- Test ETL using DO data (with full external data access)
- Verify and document QA
- Submit Achilles/DQD/AresIndexer results to central location regularly
- ETL Development Planning and Management:
- Review ETL testing and progress (TCs/meetings)
- ETL Execution:
-
Data Quality Assessment
- QA/Acceptance testing:
- Evaluate accuracy and completeness of mapping
- Review and approval by DO
- QA/Acceptance testing:
-
Documentation
- Mapping Documentation and Themis Checks
- Transformation/Technical Documentation
-
Project Management Througout
- Organization of tasks, milestones, and follow-up
R, SQL, Python, or any preferred data analysis software. Examples provided below are for R and SQL. [The Book of OHDSI Chapter 9] (https://ohdsi.github.io/TheBookOfOhdsi/SqlAndR.html) provides an overview of analysis of OHDSI data in R and SQL; note that you will not be able to avail yourselves of OHDSI software tools when analyzing your exported data for the reason explained above.
Open, rigorous and reproducible research: A practitioner’s handbook From Standord Data Science
DMP Tool: https://dmptool.org/ https://sharing.nih.gov/data-management-and-sharing-policy/planning-and-budgeting-for-data-management-and-sharing/writing-a-data-management-and-sharing-plan#after
Software Carpentry is a website that provides free online lessons to researchers wanting to enhance their programming skills for data analysis. This website offers free online lessons on a variety of useful topics including:
Additional resources:
- DataCamp
- Khan Academy
- Codecademy - Learn Python 2
- Python Data Science Handbook
- R for Data Science
- [Introduction to Programming (NIAID, NIH)](https://bioinformatics.niaid.nih.gov/resources - 70.3.1)
- [Python Programming (NIAID, NIH)](https://bioinformatics.niaid.nih.gov/resources - 70.3.2)
- [Data Analysis with Python and Pandas (NIAID, NIH)](https://bioinformatics.niaid.nih.gov/resources - 70.3.3)
- [Data Visualization with Python (NIAID, NIH)](https://bioinformatics.niaid.nih.gov/resources - 70.3.4)
- Source:NIH All of US Study
Hello! Please familiarize yourself with the following tools and resources which will help you throughout this course and your OHDSI journey.
Check out the OHDSI Forums Introduce yourself on the "Welcome to OHDSI" thread.
Bookmark The Book of OHDSI
Check out OHOP CDM FAQ
Join the OHDSI Microsoft Teams environment.
Check out the MIMIC-IV demo data set in OMOP CDM format!
Register with EHDEN Academy
Visit the Atlas Demo and Athena.
Bookmark the OHDSI YouTube tutorials and workshops
Visit the OHDSI Community Dashboard
Bookmark OMOP Common Data Model (ohdsi.github.io)
Learn about GitHub if you don't already know.
Plan to attend an OHDSI Community call
Learn about OHDSI Workgroups
Follow OHDSI on social media: Twitter LinkedIn
Subscribe to the OHDSI Newsletter
Learn about past and upcoming OHDSI events
Learn about OHDSI software
Look up individual concepts in Athena
Check out useful OHDSI-related documentation here: NIH ALL of US OMOP Documentation
Clinical Registries in OHDSI - September 2022
Matentzoglu N, Balhoff JP, Bello SM, Bizon C, BrushM, Callahan TJ et al. A Simple Standard for Sharing Ontological Mappings (SSSOM). Database. 2022. 2022:baac035, DOI: 10.1093/database/baac035.
Mapping Commons. SSSOM: Simple Standard for Sharing Ontological Mappings. Wiki [Internet]. Available from: https://mapping-commons.github.io/sssom/about.
Mapping Commons. SSSOM: Simple Standard for Sharing Ontological Mappings. GitHub [Internet]. Available from: https://github.com/mapping-commons/sssom.
https://www.w3.org/2004/02/skos/
-
July 6, 2023:
Data Quality Dashboard output demo
By: Jared Houghtaling
-
July 27, 2023:
Flowsheet follow-up
By: Polina Talapova & Jared Houghtaling
-
August 3, 2023:
OMOP Standardized Vocabularies - Part 1
By: Jared Houghtaling and Polina Talapova
-
August 17, 2023:
OMOP Standardized Vocabularies - Part 2
By: Polina Talapova
-
August 24, 2023:
How to download and set-up a DDL (Demo)
By: Jared Houghtaling
-
August 31, 2023:
Demo of WhiteRabbit and RabbitInAHat
By: Jared Houghtaling
-
September 7, 2023:
ARES usefulness for ETL at Tufts
By: Jared Houghtaling
-
September 14, 2023:
Google form introduction for site progress tracking
By: Jared Houghtaling
-
September 21, 2023:
Sample ETL Process
By: Jared Houghtaling
October 12, 2023:
Google Form for Site Progress Tracking
With Jared Houghtaling and Andrew Williams
-
October 26, 2023:
Review and Prioritization of DQD Results, and Discussion of DQD Issue Severity
With Jared Houghtaling
-
November 2, 2023:
Principles of Mapping and Vocab Gaps Identification
With Polina Talapova
-
November 9, 2023:
Usagi & STCM Demo
With Polina Talapova & Jared Houghtailing
- How racial biases in medical algorithms lead to inequities in care | PBS News Weekend
- AI Reveals its Biases by Generating What it Thinks Professors Look Like | PetaPixel
- Does “AI” stand for augmenting inequality in the era of covid-19 healthcare? | The BMJ
- The AUC Data Science | Initiative (aucenter.edu)
- ReCode-Report.pdf (data.org)
- ADDI Researcher Roundtable: The Importance of Diversity in Dementia Research on Vimeo
- Advancing Antiracism, Diversity, Equity, and Inclusion in STEMM Organizations: Beyond Broadening Participation |The National Academies Press
- Data Literacy: The Composition Effect | Education | St. Louis Fed (stlouisfed.org)
- A lecture by Pilar Ossorio at MLHC Professor of Law and Bioethics at the University of Wisconsin Law School
- This paper by Dunkelau and Leuschel that summarizes Fairness-aware Machine Learning
- Paper by Gichoya et al about minimixing bias in AI
- A conversation with Cathy O’Neil, author of the critically acclaimed Weapons of Math Destruction - YouTube
- Nicole G. Weiskopf (NYU) and Carolyn Thompson (UCSD) – Bias in EHR - YouTube
- This Obermeyer et al paper on dissecting racial bias in an algorithm used to manage the health of populations - Science, 2019
- Fair ML Keynote talk + Microsoft talk + slides - Science, 2019
- Maria Hightower, M.D., M.B.A., MPH Chief Digital Technology officer of the University of Chicago Medicine comments on the racial bias in AI and the algorithm described above - Science, 2019
- How scientists are subtracting race from medical risk calculators - Science, 2021
- Race After Technology, by Ruha Benjamin [Professor of African American studies at Princeton University], summarizes how technology [from data collection, data imputation, government policy, etc] can play a role in different outcomes in society - Science
This is the main REDCap training page: REDCap Resources
From University of Colorado, below are excellent REDCap resources: University of Colorado REDCap Resources
- Background Information: Learn more about the background of REDCap at UC Denver, general policies for use, and some REDCap best practices (video)
- COMIRB Template (user guide)
- Best Practices (user guide)
- 18 HIPAA Identifiers & PHI (University of Maryland) (user guide)
- Login and Project Creation: Learn how to log in to your REDCap account and create a new project (video)
- Project Setup Page: Explore the project home, project setup, other functionality, and project revision pages. Learn how to customize the settings for your projects, set user rights, use data access groups, and move your project to Production (video)
- Online Designer: Learn about the basic steps in designing your project. Learn how to set up forms; create a new field; validate fields; code multiple choice questions; use yes/no and true/false fields; create slider fields; and how to use file upload fields, descriptive fields, and section headers (video)
- Online Designer - Advanced Features: Learn how to use advanced features in the Online Designer such as branching logic, calculated fields, matrices, piping, and the data dictionary. Review some best practices in form creation (video)
- Longitudinal Database: Learn about the longitudinal data collection set up and how it varies from a standard or classic data collection instrument. Learn how to define events and assign forms to specific events, use the scheduling module, and advanced calendar features available only in longitudinal studies (video)
- Repeating Forms: Learn how to use repeating forms and events (video)
- Action Tags: Learn how to use the most popular and powerful action tags (video)
- Action Tags List: List of all action tags with descriptions (user guide)
- Alerts and Notifications: Learn about the alerts and notifications feature (video)
- Calculations: Learn how to use simple calculations, if/then statements, nest calculations, and datediff calculations (video)
- Field Embedding: Explore how to use the field embedding feature to format data entry and survey forms (video)
- Formatting REDCap Forms with HTML (user guide)
- Logic Guide: Learn how to format logic in REDCap (user guide)
- Smart Variables: Learn how to use smart variables in REDCap (video)
- Data Entry: Learn how to add new records, edit existing records, use advanced data entry features, and navigate between records (video)
- Missing Data: Set specific missing data codes for your entire project (video)
- Advanced Features: Learn how to use some advanced REDCap features, including the randomization feature, the logging feature, the field comment log, the data import tool, and the data quality tool (video)
- Data Imports: A detailed look at use the import tool to import data from another source (video)
- Randomization: Learn how to use the randomization feature (video)
- Reports and Exports: Learn how to create a report, review basic statistics and charts, and export a report or full data set (video)
- Public Reports: Learn how to make reports accessible with a public link (video)
- Surveys: Learn how to enable surveys, the benefits of using surveys, setting up your survey, and the different ways you can distribute both initial and follow-up surveys (video)
- Survey Login: Learn how to use the survey login feature (video)
- Survey Queue: A deep dive into using the survey queue (video)
- PDF Auto-Archiver and e-Consent: Use REDCap's e-Consent framework to consent participants and save PDFs of the consent (video)
- Twilio: Learn how to use Twilio to send your survey invitations via text message or allow participants to take the survey via voice call (video)
- Twilio Guide (user guide)
- Double Data Entry: Learn how to use REDCap's Double Data Entry module (video)
- Mobile App Guide (user guide)
- Multi-Language Management Manual (user guide)
We hold a REDCap user group meeting for our active users after every major upgrade twice a year to discuss the new features. You can watch the recordings of our previous group videos on Vimeo:
- User Group Meeting 2/23/2023: MyCap integration, automated invitations with repeating surveys, file repository improvements
- User Group Meeting 7/19/2022: Multi-language management updates, survey start time and survey duration, FAQ updates
- User Group Meeting 3/9/2022: Multi-language management, public reports, @IF form display logic
- User Group Meeting 8/11/2021: Project dashboards, smart variables, smart charts, and smart tables
- User Group Meeting 3/31/2021: @CALCDATE/@CALCTEXT and special functions
- User Group Meeting 8/26/2020: eConsent, field embedding
- Can I add users to my project? Anyone creating projects should attend a tutorial (see schedule on Tutorials page) which focuses on project creation and REDCap use policies. If you have attended a
tutorial you can request limited accounts for others who will not be designing/managing projects but only doing data entry, data export, etc. To request an account email the REDCap administrator with the person's name and work email address. By doing this you are also taking responsibility for training them.
-
Why is the Participant ID the first field in the project? When you set up a REDCap database your record identifier e.g. Participant ID must be the first field on the first form so that REDCap will link all following data on all forms for that record. There is no need to repeat the record identifier in each form.
-
How do I code categorical variables? If your categorical variable has numeric response options be sure to assign a value that is the same number to avoid confusion in analysis. For example if the question is "How many times did you ...." and the options are 0,1,2,3,4,5 you should assign values of 0-5 (note: you cannot use the auto-assign feature of REDCap because it will start with 1).
-
When should I use the dropdown field type? Use dropdown field types instead of radio buttons for categorical variables on your data entry forms. REDCap allows you to type the first character of a label to select that option in a dropdown which is much easier than having to individually select each radio button with your mouse.
-
How do I tell the difference between a radio and checkbox field? There is a visual cue to tell you whether a field is a radio button (single option) or a checkbox (choose all that apply). Radio buttons are round, checkboxes are square.
-
How do I format branching logic or a calculation with checkbox fields? Checkbox or "choose all that apply" fields are coded slightly differently from other categorical fields such as radio or dropdown. In those each option is set to equal a unique value e.g. 1=red, 2=blue, 3=green. Because any or all of the checkbox field options can be selected, each option is treated as a separate field that is either checked or unchecked (coded 1 or 0). In your exported dataset you will see that each option has become a separate variable with the number of the option as part of the variable name e.g. color(1), color(2), color(3). When using options from a checkbox field in a calculation or in branching logic instead of writing "color = 3" for example you need to write "color(3)=1" meaning that option 3 of the variable "color" has been selected.
-
How do I hide a section header field type using branching logic? Section header fields follow the branching logic for all fields until the next section header so to hide a section header all fields until the next section header must also be hidden.
-
How do I format greater than/less than conditions in my branching logic? When using a greater than/less than (><) condition in branching logic don't put quotation marks around the value as you normally would when using equal to.
-
How do I test branching logic and calculated fields in my form or survey? To test branching logic or calculated fields enter a test record into your database or survey. These functions do not work on the Preview screen. You can remove all test records when you move to production.
-
How do I add an “other” option in a multiple field question that a participant can write in their answer? To include an "Other" option in a multiple choice question that will allow respondents to write in an answer, add a text field that is only displayed when the Other option is selected. If you want it to appear right next to the “Other” option in the multiple choice question you’ll need to use field embedding. You’ll follow the step above, but then additionally you’ll need to tell REDCap to embed the field with the multiple choice answers—this is done by putting the variable name in { } curly brackets where you want it to appear.
-
Can I base a calculated field off another calculated field? If you are using calculated fields, avoid creating second-level calculations, i.e. using the results of a calculation as part of another calculation. These fields will not reliably calculate - even though values may appear in the field onscreen, the field may be blank in your exported dataset. Keep in mind the general recommendation to do calculations as part of analysis and keep only raw data in your REDCap database.
-
Why isn’t my calculation saved? If you add a calculated field after you have collected the data used in the calculation you will need to resave the form containing the calculation to trigger REDCap to perform the calculation and populate the field. To update them all at once, you can go to Data Quality under Applications on the sidebar and run Rule H. It will show all fields where the calculated values don’t match what is saved and give you the option to update them all.
-
Can I display an image in my form or survey? To display an image on your data entry form or survey use the descriptive text field type which has a field upload feature. For more information please see Formatting REDCap Forms with HTML.
-
Can I customize the appearance of my form or survey? REDCap allows some customization of form appearance using HTML code. These include font size, font color, and spacing/indentation of field label text.
-
How do I set up the event grid in a longitudinal project? When setting up a Longitudinal project event grid, if you are not using the Scheduling module, you don't need to set specific "days offset" but you still need to enter something to tell REDCap the order of your events. If you leave all zero's REDCap will put your events in alphabetical order. So you can just put 1, 2, 3, etc. You may also want to use increments of 5's in case you later need to insert a new event.
-
What happens if I accidently delete an event? In a Longitudinal Model database if you accidentally delete an event your data will not be lost just hidden. When you restore the event with the assigned forms the data will also be restored.
-
Can I change the coding of my fields while in production? Although you can make changes to your project fields after moving your project to Production, keep in mind that changes to coding for categorical fields will impact your existing data. For example if you have a field with Yes/No responses that are coded 10 and you change these to 21 then what was originally coded as Yes will now be No - since you have changed the meaning of the value 1.
-
How do I test my database before moving to production? It's a good idea to test your database or survey before moving to Production by entering a few records of either real or fake data. When moving to Production you can choose to keep or delete these records.
-
How do I review the changes I have made before submitting? Before submitting post-production changes for review, you can see whether they will cause any problems by selecting the "view a detailed summary of all drafted changes" link located next to the Submit Changes for Review button.
-
Will I lose data if submitting changes while in production? Project changes made after moving to production must be reviewed prior to being implemented to reduce the risk of data corruption due to a change. However, if you make a change that cannot possibly impact existing data (e.g. create a new field), once you submit the change for approval it will be approved automatically - you won't have to wait for manual review and approval by the administrator. To view whether you changes create potential issues while you're in draft mode, go to the "view a detailed summary of all drafted changes" link.
-
What should I do if I need to make significant changes while in production? If your project is in production and you need to make several changes, consider making a copy of the project (which will be in development), making the changes there, then using the data dictionary to implement the changes all at once in the production project. Alternatively, you can ask the REDCap administrator to move your project back to development.
-
How do I combine my instruments into a single survey? If you have multiple instruments that you want to send out as a single survey, combine them onto a single REDCap form. You can insert page breaks between instruments using the Section Header field type.
-
How do I hide calculated fields in a survey? If you are including survey responses in a calculation but don't want the respondent to see the calculations, create a separate data entry form and put the calculation fields there. The calculations will be triggered when the survey is submitted.
-
How do I schedule a survey to be sent at a specific date/time? Using the "automated invitations" feature you can schedule survey to be sent at a specific date/time as well as based on a specific response to a previous form or survey. See detailed instructions on how to do this in the Help/FAQ page.
-
How can I set up surveys to go out automatically? If you are using online surveys, you can schedule them to be sent automatically at certain dates or based on specified conditions being met. See Automated Invitations in the Online Designer for instructions.
-
How do I personalize an email invitation? REDCap allows you to customize field labels or survey invitations using "piping". This means you can insert the response to one field e.g. first name into the label of another field or into a survey invitation text. To do this just put the variable name in square brackets where you want the customized text. For example if the
respondent's first name is in a variable called "fname" you can add it to the label of another field like this: [fname] what is your favorite color? Similarly, when you write a survey invitation you can use: Dear [fname], please complete the attached survey. Whatever name has been entered in the field fname will appear in place of the variable name.
-
How do I track survey responses? You can track who has responded to a survey by using the Participant List option. In addition, you can identify individual responses using the Participant Identifier feature. Both of these options are found in the Manage Survey Participants section of your project.
-
Can I add or delete a survey response? It is possible to edit or delete survey responses. To do this, check the Edit Surveys option in the User Rights section.
-
How do I verify if a survey respondent changed their answers? If you are concerned that survey respondents changed their answers before submitting their survey e.g. to try to qualify for a study you can check their responses in the Logging Tool.
-
What is the logging tool? You can use the logging tool to troubleshoot issues that arise that may be due to a change in a data value, calculations and branching logic no longer working, etc. In the log you can filter by record, user, and event type.
## Analysis with SQL (OHDSI/OMOP)
The [OMOP Query Library](https://data.ohdsi.org/QueryLibrary/) is a library of commonly-used SQL queries for the OMOP Common Data Model (CDM).
## Analysis with R
Below are some sample R queries that demonstrate how to read in OMOP tables from CSV files, join them based on the `person_id` and `visit_occurrence_id` fields, and search for specific criteria.
Note: Adjust the file paths and column names accordingly based on the actual structure and location of your CSV files. The queries below are a generic representation and may need adjustments based on the specifics of your data set.
### Reading CSV files into R data frames:
```R
# Read the CSV files into R data frames
person_df <- read.csv("path_to_person_table.csv", header=TRUE, stringsAsFactors=FALSE)
visit_occurrence_df <- read.csv("path_to_visit_occurrence_table.csv", header=TRUE, stringsAsFactors=FALSE)
condition_occurrence_df <- read.csv("path_to_condition_occurrence_table.csv", header=TRUE, stringsAsFactors=FALSE)
Join tables based on person_id:
When a person has multiple visits in the visit_occurrence table, joining the person table with the visit_occurrence table will result in multiple rows for that person, each corresponding to a different visit. This is a standard one-to-many join operation.
## Join person with visit_occurrence on 'person_id'
person_visit_df <- merge(person_df, visit_occurrence_df, by="person_id")# Join the person-visit result with condition_occurrence on both 'person_id' and 'visit_occurrence_id'
full_df <- merge(person_visit_df, condition_occurrence_df, by=c("person_id", "visit_occurrence_id"))# Define a list of person_ids to search for
search_person_ids <- c(1, 2, 3, 4, 5)
# Filter the data frame to only include rows with person_ids in the list
filtered_by_person_df <- subset(full_df, person_id %in% search_person_ids)# Define a specific condition concept code to search for
search_condition_concept_id <- 1234567
# Filter the data frame to only include rows with the specified condition concept code
filtered_by_condition_df <- subset(full_df, condition_concept_id == search_condition_concept_id)# Define a date range to search for
start_date <- as.Date("2020-01-01")
end_date <- as.Date("2020-12-31")
## Filter the data frame to only include rows within the date range
filtered_by_date_df <- subset(full_df, visit_start_date >= start_date & visit_start_date <= end_date)