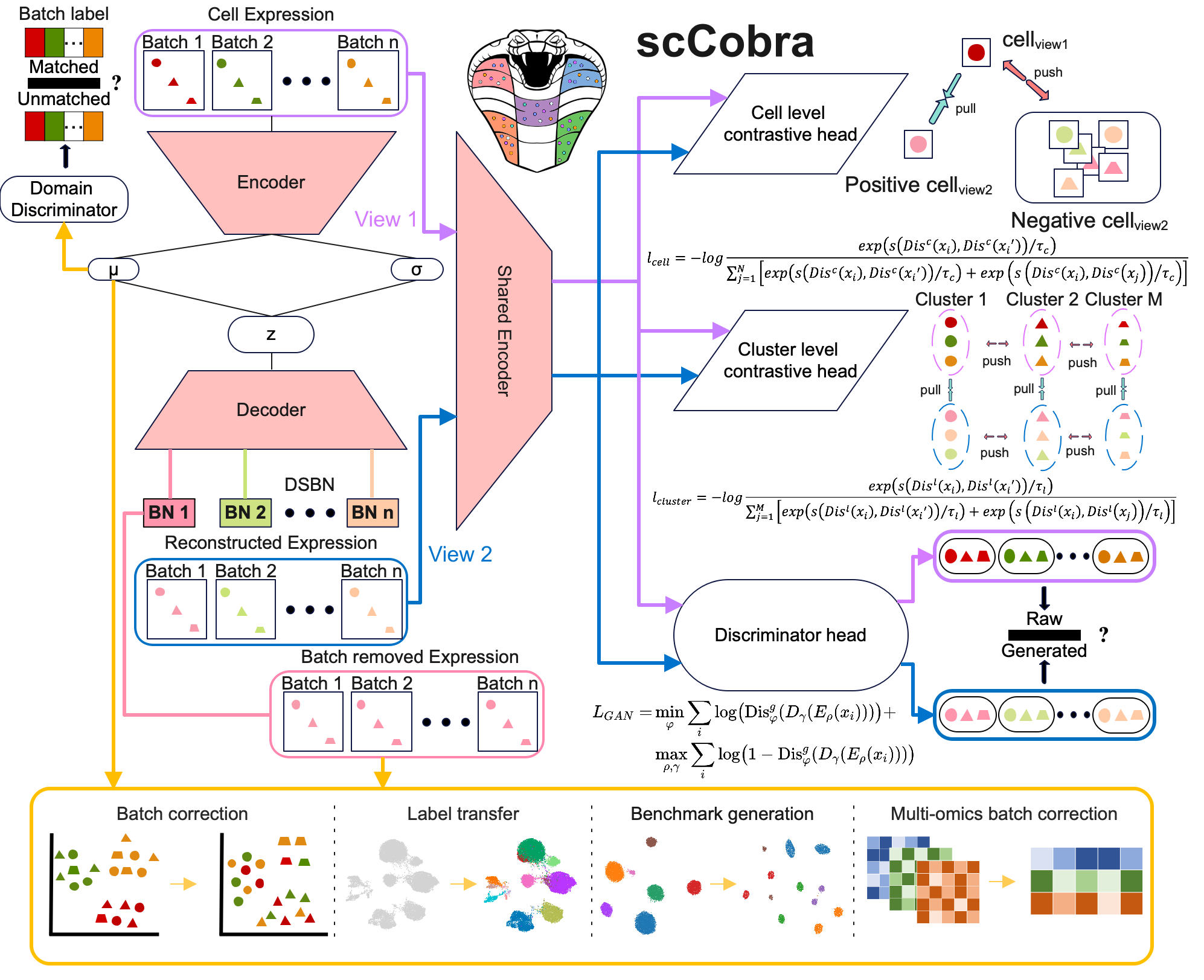
scCobra: Contrastive cell embedding learning with domain adaptation for single-cell data integration
scCobra is designed for integrating single-cell data from different batches and/or sequencing platforms. scCobra is a two-phase model. In phase 1, we let the original data enter the random dropout layer twice independently, so each cell will get two augmented views. Then a weight-sharing encoder is employed to encode the cell inputs in two augmented views into reduced latent embeddings. Here we employ a set of domain-specific batch normalization layers to normalize the embeddings from various sources (experimental batches). Following that, the projection head maps the latent cell embeddings nonlinearly to the same joint reduced space (h) for contrastive learning. In phase 2, we used the generative adversarial networks (GAN) to fuse different experiment batches of cells. Specifically, we use an adversarial training strategy to alternately train the discriminator and encoder, aligning the gene expression distribution of datasets from different experimental batches. After training, we use the trained encoder to encode cells directly, and the resulting cell embeddings (z) will be used for downstream tasks, such as cell clustering or trajectory inference of the integrated dataset. The scCobra method demonstrates superior performance as benchmarked with several other state-of-the-art methods. Please refer to our manuscript for details.
Step 1: Create a conda environment for scCobra
# Recommend you to use python above 3.9
conda create -n scCobra conda-forge::python=3.9 bioconda::bioconductor-singlecellexperiment=1.20.0 conda-forge::r-seuratobject=4.1.3 conda-forge::r-seurat=4.3.0 bioconda::anndata2ri=1.1 conda-forge::rpy2=3.5.2 bioconda::r-signac bioconda::bioconductor-ensdb.hsapiens.v75 bioconda::bioconductor-biovizbaseconda-forge::r-irkernel conda-forge::ipykernel
# Install scanpy scib episcanpy snapatac2
pip install scanpy scib episcanpy snapatac2
# You can install addtional packages: https://scib.readthedocs.io/en/latest/index.html
# Install pytorch
pip3 install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118Step 2: Clone This Repo
git clone https://github.com/GlancerZ/scCobra.gitYou can click the dataset name to download
- simulated dataset contains 12097 cells with 9979 genes, has 7 cell types from 6 batches
- pancreas dataset contains 16382 cells with 19093 genes, has 14 cell types from 9 batches
- Lung atlas dataset contains 32472 cells with 15148 genes, has 17 cell types from 16 batches
scCobra is jointly developed by Bowen Zhao and Yi Xiong from Shanghai Jiaotong University and Jun Ding from McGill University.