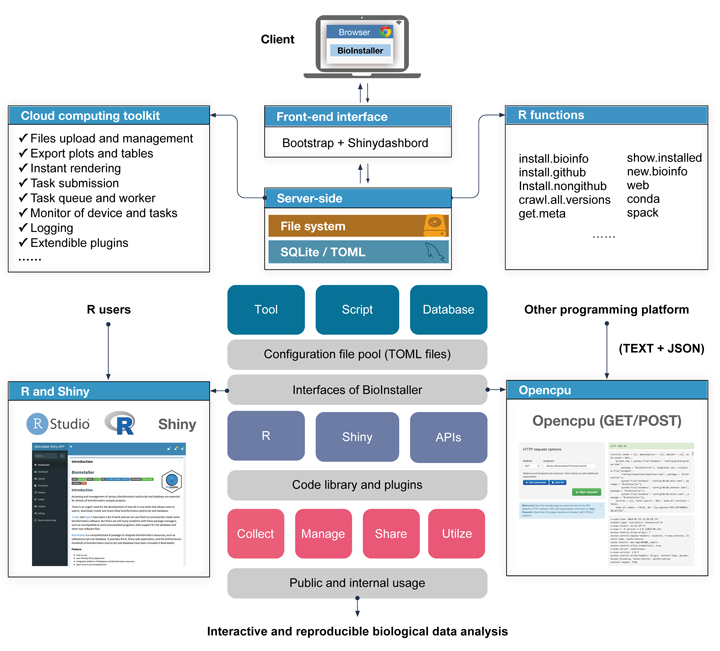
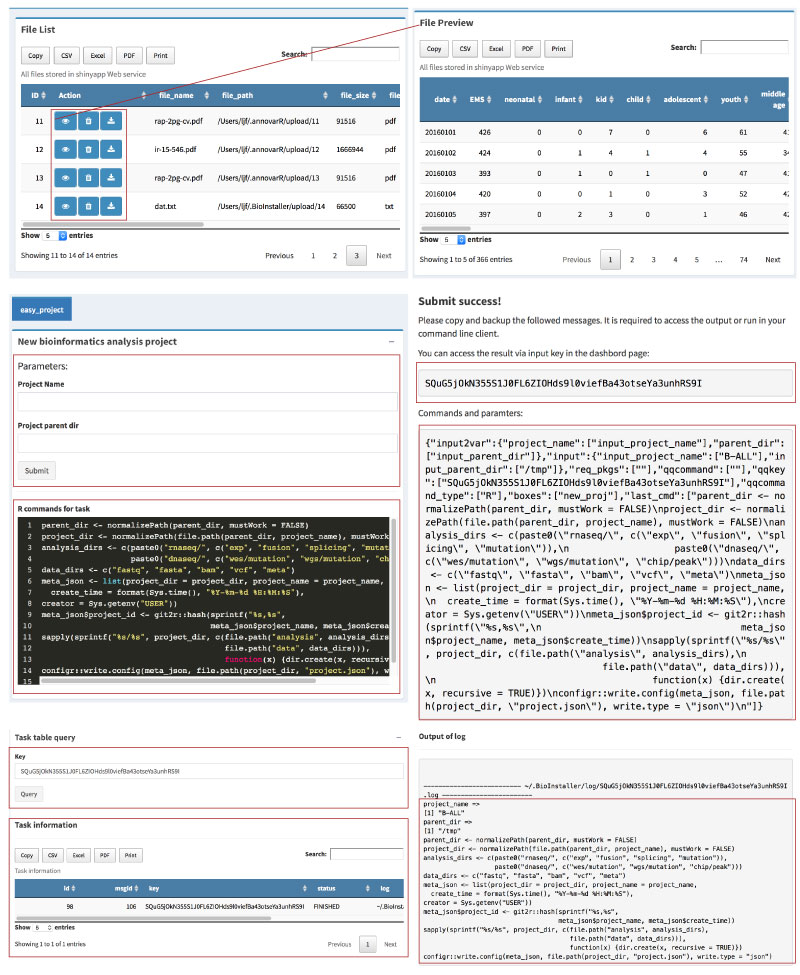
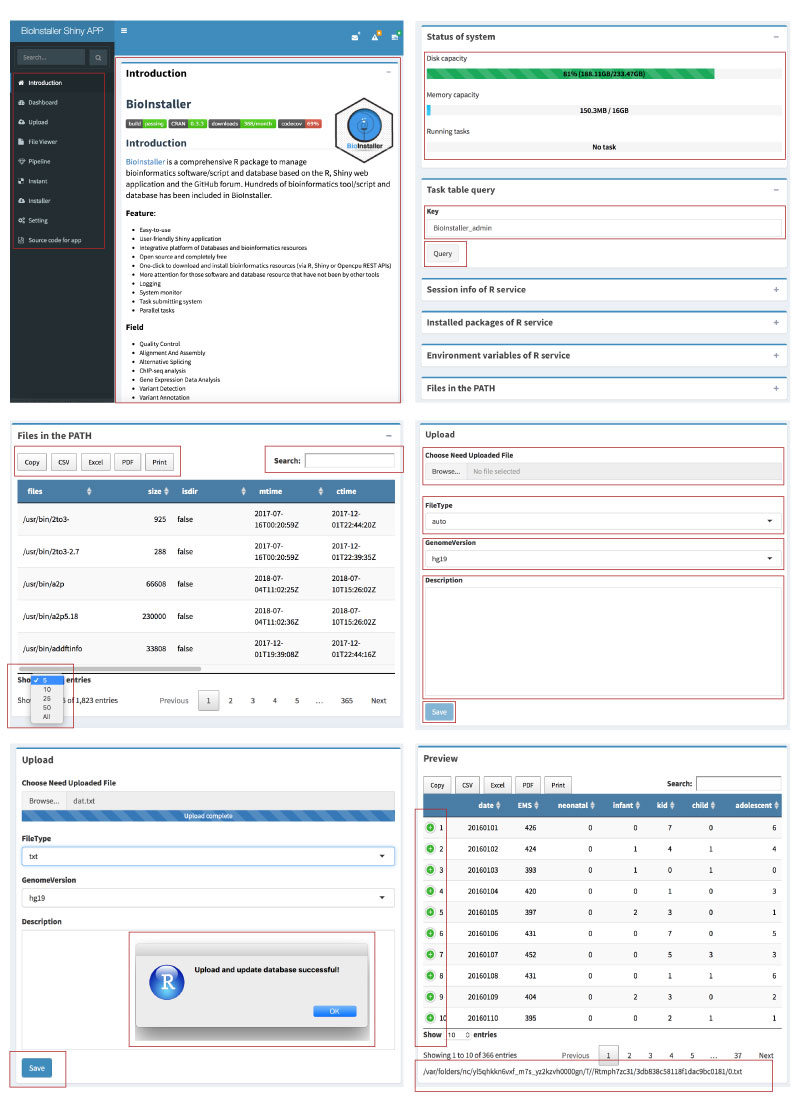
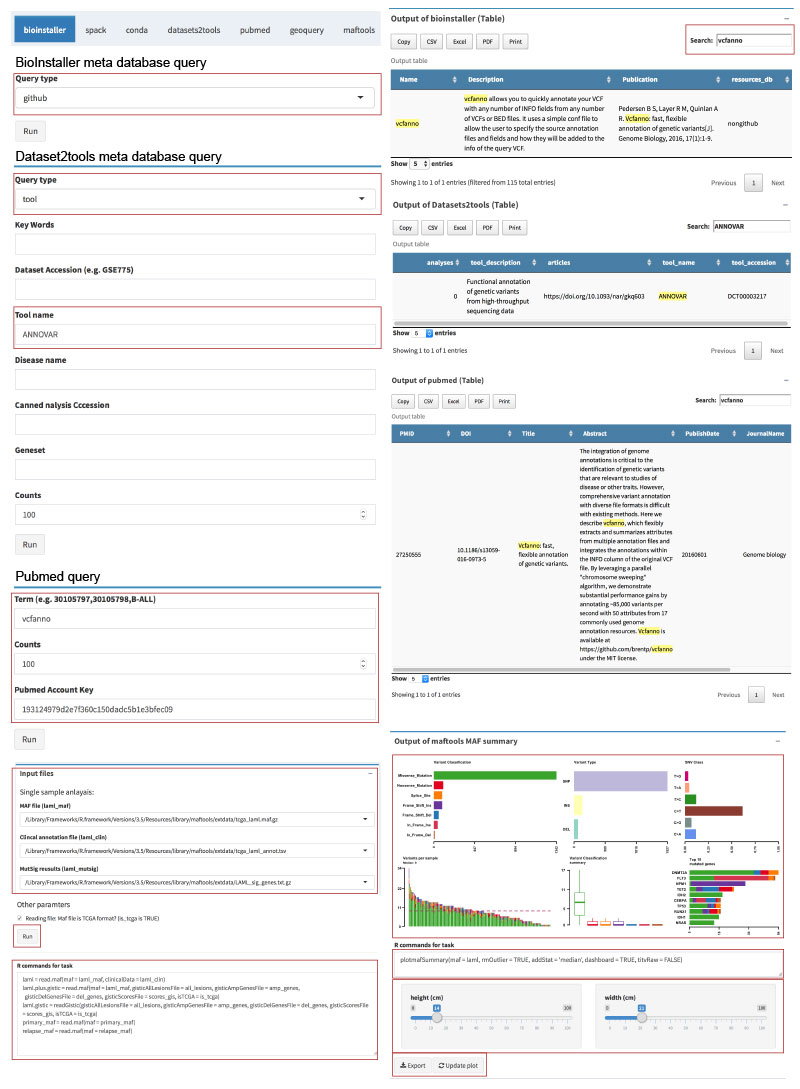
Accessing and management of various bioinformatics tool/script and database are essential for almost all bioinformatics analysis projects.
There is an urgent need for the development of new all-in-one tools that allows users to search, download, install and share these bioinformatics tool/script and database.
Conda and Spack have done a lot of work and we can use them to conveniently install some bioinformatics software. But there are still many problems with these package managers, such as incompatible to some precompiled programs, little support for the database and other non-software files.
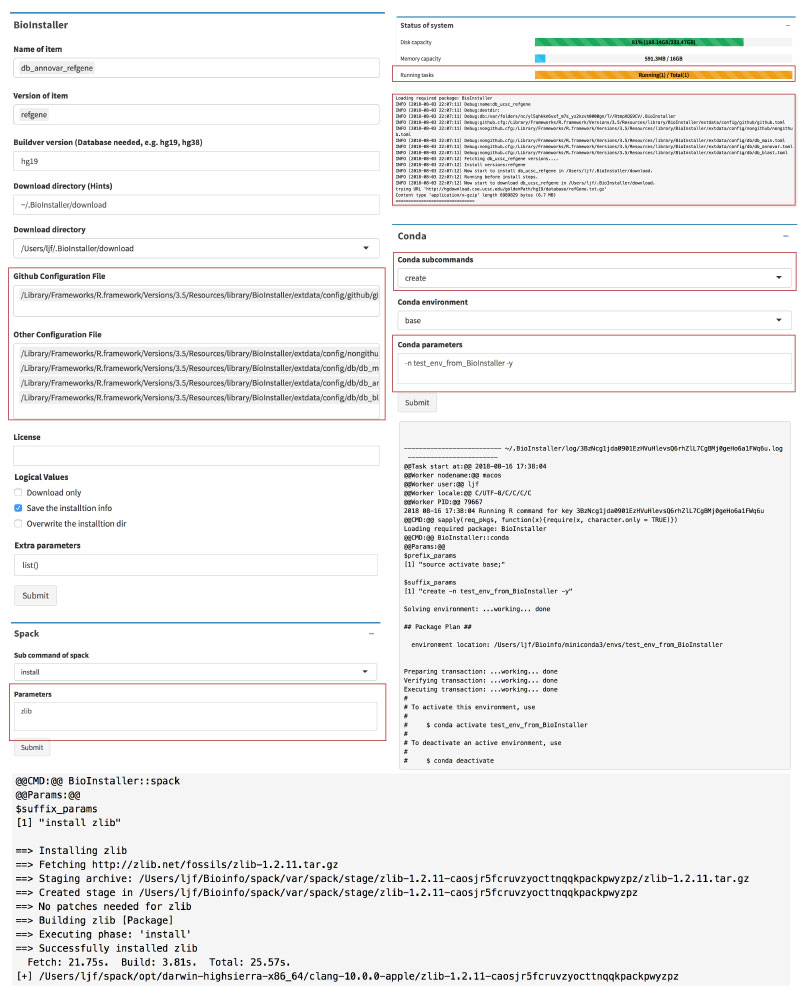
BioInstaller is a comprehensive R package to integrate bioinformatics resources, such as software/script and database. It provides the R, Shiny web application, and the GitHub forum. Hundreds of bioinformatics tool/script and database have been included in BioInstaller.
Feature:
- Easy-to-use
- User-friendly Shiny application
- Integrative platform of Databases and bioinformatics resources
- Open source and completely free
- One-click to download and install bioinformatics resources (via R, Shiny or Opencpu REST APIs)
- More attention for those software and database resource that have not been by other tools
- Logging
- System monitor
- Task submitting system
- Parallel tasks
Field
- Quality Control
- Alignment And Assembly
- Alternative Splicing
- ChIP-seq analysis
- Gene Expression Data Analysis
- Variant Detection
- Variant Annotation
- Virus Related
- Statistical and Visualization
- Noncoding RNA Related Database
- Cancer Genomics Database
- Regulator Related Database
- eQTL Related Database
- Clinical Annotation
- Drugs Database
- Proteomic Database
- Software Dependence Database
- ......
# install the latest developmental version
# than start the BioInstaller R Shiny application
# the document is still under construction
BioInstaller::web()
#You can install this package directly from CRAN by running (from within R):
install.packages('BioInstaller')# install.packages("devtools")
devtools::install_github("JhuangLab/BioInstaller")- GitHub resource
- GitHub resource meta information
- Non GitHub resource
- Non Github resource meta infrmation
- Database
- Web Service
- Docker
Quality Control:
- FastQC, PRINSEQ, SolexaQA, FASTX-Toolkit ...
Alignment and Assembly:
- BWA, STAR, TMAP, Bowtie, Bowtie2, tophat2, hisat2, GMAP-GSNAP, ABySS, SSAHA2, Velvet, Edean, Trinity, oases, RUM, MapSplice2, NovoAlign ...
Variant Detection:
- GATK, Mutect, VarScan2, FreeBayes, LoFreq, TVC, SomaticSniper, Pindel, Delly, BreakDancer, FusionCatcher, Genome STRiP, CNVnator, CNVkit, SpeedSeq ...
Variant Annotation:
- ANNOVAR, SnpEff, VEP, oncotator ...
Utils:
- htslib, samtools, bcftools, bedtools, bamtools, vcftools, sratools, picard, HTSeq, seqtk, UCSC Utils(blat, liftOver), bamUtil, jvarkit, bcl2fastq2, fastq_tools ...
Genome:
- hisat2_reffa, ucsc_reffa, ensemble_reffa ...
Others:
- sparsehash, SQLite, pigz, lzo, lzop, bzip2, zlib, armadillo, pxz, ROOT, curl, xz, pcre, R, gatk_bundle, ImageJ, igraph ...
Databases:
- ANNOVAR, blast, CSCD, GATK_Bundle, biosystems, civic, denovo_db, dgidb, diseaseenhancer, drugbank, ecodrug, expression_atlas, funcoup, gtex, hpo, inbiomap, interpro, medreaders, mndr, msdd, omim, pancanqtl, proteinatlas, remap2, rsnp3, seecancer, srnanalyzer, superdrug2, tumorfusions, varcards ...
You can use the BioInstaller in Docker since v0.3.0.
docker pull bioinstaller/bioinstaller:develop
docker run -it -v /tmp/download:/tmp/download bioinstaller/bioinstaller:develop RPlease fork the GitHub BioInstaller
repository, modify it, and
submit a pull request to us. Especialy, the files list in contributed section should be modified when you see a tool or database that not be
included in the other software warehouse.
R package:
Related Other Resources
Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License