-
-
Notifications
You must be signed in to change notification settings - Fork 4
Essentials

The MIMBCD-UI project is a project that deals with several modalities of medical images. Our Framework will incorporate information from following (see next Figure) different modalities: (i) MammoGraphic (MG) images (both views CC and MLO); (ii) UltraSound (US) images; (iii) Magnetic Resonance Imaging (MRI) volumes; and (iv) Text. If you have some questions, please follow our FAQ page.

In this page, it is intended to be used as a reference for the concepts of the project Framework. If you are new to our project, we recommend that you start by reviewing the concepts below and then looking at the various prototypes to see how to use each.
Git is an indispensable tool within the MIMBCD-UI initiative, enabling researchers and developers to collaborate effectively on complex codebases. In a project where the integrity and reproducibility of medical imaging experiments are paramount, the Git tools allows the team to manage and track every change meticulously. Researchers can create branches to explore new ideas or test novel algorithms, ensuring that experimental changes do not disrupt the main codebase. Once validated, these changes can be seamlessly merged back, preserving a comprehensive history of the project’s evolution and maintaining the scientific rigor required in medical research.
For the MIMBCD-UI initiative, GitHub is more than just a repository hosting service—it’s a vital platform for collaboration across multiple research institutions. Given the interdisciplinary nature of the project, which spans medical imaging, computer science, and human-computer interaction, GitHub provides the necessary tools to manage version control, coordinate tasks, and document progress. Features like pull requests allow team members to review and discuss code changes before they are integrated, ensuring that all contributions meet the project’s high standards. Issue tracking and discussions facilitate transparent communication, making it easier to manage the complexities of a large-scale research project spread across different labs and countries.
Within the MIMBCD-UI initiative, commit messages are not just technical notes, they are a critical part of the project’s documentation. Each commit message captures the details of changes made, providing a clear narrative that explains why certain decisions were taken. In a research project where every modification can impact experimental outcomes and subsequent analysis, these messages are invaluable. They ensure that all team members can trace the rationale behind changes, which is crucial for maintaining continuity, especially when revisiting past work or when onboarding new collaborators.
In the context of the MIMBCD-UI initiative, using a consistent and structured tagging system in commit messages is essential. These tags allow team members to quickly understand the purpose and scope of each commit, whether it’s adding new functionality, fixing bugs, or updating documentation. Given the project’s scale and the variety of disciplines involved, clear tags help streamline collaboration by making it easier to navigate the commit history and ensure that all contributions are aligned with the project’s goals.
-
[NEW]:
- Use for: Introducing completely new elements, such as modules, systems, or major features.
-
Example:
[NEW] Implement OAuth2.0 authentication
-
[CREATE]:
- Use for: Initial implementation of new files, directories, or basic features.
-
Example:
[CREATE] Initial implementation of user authentication module
-
[ADD]:
- Use for: Adding new components, code, or assets to an existing structure.
-
Example:
[ADD] Include new icons for user interface
-
[FEATURE]:
- Use for: Introducing a new feature to the codebase.
-
Example:
[FEATURE] Add multi-language support to the application
-
[UPDATE]:
- Use for: Modifying existing features, improving code, or making enhancements.
-
Example:
[UPDATE] Refactor payment processing logic for improved efficiency
-
[FIX]:
- Use for: Bug fixes or any corrections to existing code.
-
Example:
[FIX] Resolve issue with date parsing in report generation
-
[REFACTOR]:
- Use for: Restructuring code without changing its functionality, typically for improving readability, maintainability, or performance.
-
Example:
[REFACTOR] Simplify database query logic in user service
-
[PERF]:
- Use for: Performance improvements or optimizations.
-
Example:
[PERF] Optimize image loading for faster rendering
-
[SECURITY]:
- Use for: Security-related changes, such as vulnerability fixes or adding security measures.
-
Example:
[SECURITY] Implement input sanitization to prevent SQL injection
-
[STYLE]:
- Use for: Non-functional changes related to code formatting.
-
Example:
[STYLE] Apply consistent code formatting across all files
-
[CONFIG]:
- Use for: Changes to configuration files, CI/CD pipelines, environment settings, or build scripts.
-
Example:
[CONFIG] Update Dockerfile for better caching during builds
-
[DOCS]:
- Use for: Documentation-related changes, such as updating the README or inline comments.
-
Example:
[DOCS] Add usage examples to the API documentation
-
[REMOVE]:
- Use for: Removing code, features, or functionality that are no longer needed.
-
Example:
[REMOVE] Eliminate redundant error logging in service layer
-
[DELETE]:
- Use for: Deleting specific files, code, or assets from the project.
-
Example:
[DELETE] Remove outdated config files from the project
Using these tags ensures that everyone in the project can quickly understand the purpose and context of a commit, which is especially useful in large, complex projects like MIMBCD-UI. They help maintain clarity and organization in the commit history, making it easier for team members to track progress, identify specific changes, and collaborate effectively across different research areas. This level of organization is crucial for ensuring that the project remains cohesive and that all contributions are aligned with the initiative’s overarching goals.
Choosing the correct tag can sometimes be tricky, especially when tags have overlapping meanings. The choice should reflect the true nature of the changes being made to ensure clarity in the commit history.
-
[NEW] vs. [CREATE]:
- [NEW] should be used when introducing something entirely new, like a module, system, or major feature. It’s typically for larger or more significant additions to the project.
- [CREATE] is best used for initial implementations, like setting up a new file, directory, or basic feature that’s part of a larger system.
-
[ADD] vs. [NEW]:
- [ADD] is for smaller additions to existing structures, such as adding new methods to a class, new icons to an interface, or additional documentation.
- [NEW] should be used when what you’re adding represents a new major component or system within the project.
-
[REMOVE] vs. [DELETE]:
- [REMOVE] can be used when removing functionality, code, or features that aren’t necessarily file-based. It’s often broader and can apply to any part of the codebase.
- [DELETE] is specifically for removing files or assets from the project. Use this tag when the action is focused on physical deletions.
-
[UPDATE] vs. [FEATURE]:
- [UPDATE] should be used for modifications or improvements to existing features or code.
- [FEATURE] is for when you’re adding a new feature to the codebase, even if it’s an extension of an existing feature set.
-
[FIX] vs. [UPDATE]:
- [FIX] is specifically for bug fixes or corrections to existing code.
- [UPDATE] is broader and includes enhancements or changes that don’t necessarily involve fixing a bug.
Understanding the nuances between these tags helps ensure that your commit messages accurately convey the nature of the changes, which is essential for maintaining a clear and navigable commit history. In a multidisciplinary project like MIMBCD-UI, where various teams and experts contribute, this clarity prevents misunderstandings and facilitates smoother collaboration. It also aids in the peer review process, allowing others to quickly grasp the significance and impact of each change, ultimately supporting the project’s scientific integrity and progress.
There are a few pre-defined settings files that you can find in config/ folders. These are JSON files and all settings can be accessed as showed below. Our User Interface (UI) settings are also available on several folders and files.
As a DICOM server, we are using the Orthanc server. Configuring Orthanc simply consists of a configuration file. Orthanc has numerous configuration that is documented in the default configuration file. Please follow the official configuration documentation for more information.
Our prototypes configuration files can be found typically on REPOSITORY_NAME/config/ folder. Also, each NodeJS repository has a package.json file. This file contains a lot of meta-data about the respective repository. Mostly, it will be used for managing dependencies of the repository and scripts. The scripts will help in generating builds, running tests and other stuff in regards to the respective repository.
Environment files are useful when deploying on several server stages. For instance, if we want to have specific values across multiple environments (e.g., prod, test, stage or dev). Our values change frequently and are highly dynamic, therefore, the environment variables can be changed easily. In our solutions, we use the env.json file on the config/ folder to manage our environments.
The following example shows us a possible env.json file sample:
{
"environment": "sample.json"
}
There are several different ways to set environment variables. Each has a slightly different use case. The dicomServer tag configures the settings regarding the DICOM server. The mainserver tag configures the settings regarding the prototype server configurations.
The following example shows us a possible sample.json file sample:
{
"dicomServer":
[
{
"prefix": "://",
"transferProtocol": "http",
"hostname": "localhost",
"portEscape": ":",
"port": "8042",
"suffix": "/",
"serviceProtocol": "wadouri"
}
],
"mainServer":
[
{
"prefix": "://",
"transferProtocol": "http",
"hostname": "localhost",
"portEscape": ":",
"port": "8080",
"suffix": "/",
"hotjar": "123456"
}
]
}
Starting with our several repositories, a User Interface (UI) is provided to use each prototype on the client-side and server-side. Simply follow the README.md information to setup each prototype on your local machine, or in a remote server. In our prototypes, we follow a Web Access to DICOM Objects by URI (WADO-URI) requests. The WADO-URI requests are HTTP requests that use the GET method as defined in IETF RFC2616. A Medium.com story, titled as "Using CornerstoneJS and Orthanc to Support Deep Learning Projects", introduces the meta tag structures but we still we summarize it here.

Each .dcm file (DICOM) contains a file header portion. We call it File Meta Information portion working as a single SOP Instance (i.e., a hierarchy of identifiers for each individual). For more information regarding your SOP Instance questions, follow this post. The header is made up of a 128-byte preamble, followed by the characters DICOM, all uppercase. The preamble must contain all zeroes if it is not used (sometimes applications will use it for proprietary data).
Usually, we use a client-server model to express our distributed structure, by partitioning tasks or workloads between the providers of a resource or service. For users accustomed to working in high-performance environments, it is natural to question whether workstations and servers have what it takes to manage medical image distribution throughout clinical institutions. For this reason, we focus on a client-server model ensuring to meet or exceed the demands of today’s radiologic environments.
Our prototypes are mainly developed on web technologies such as HTML5, CSS3 and JavaScript. Like web applications, they provide a platform-independent solution to display DICOM studies for all medical modalities. The fully customizable application is a powerful collection of JavaScript and NodeJS libraries, as well as web services, while it is perfect for any integrator who needs a fast, lightweight Medical Imaging (MI) solution (see next Figure) without sacrificing any features that health care professionals demand, including 3D volumes.

Each prototype takes full advantage of everything modern web browsers offer, giving us a shorter learning curve and confidence that such applications will satisfy clinician's demands for User eXperience (UX) and usability. The plethora of fast access, client-side tools, and medical-specific image processing make it a premier choice for working in the health care space on a research project. The shift to a web-based system for image assessments rather than a workstation-based installed application will improve workflow efficiency. Also, it will enhance accessibility and promote collaborative image review for the radiologists at cooperating clinical institutions (see next Figure). To achieve our design goals, the viewer and all of its functionality will be delivered to client-side exclusively through the web browser.
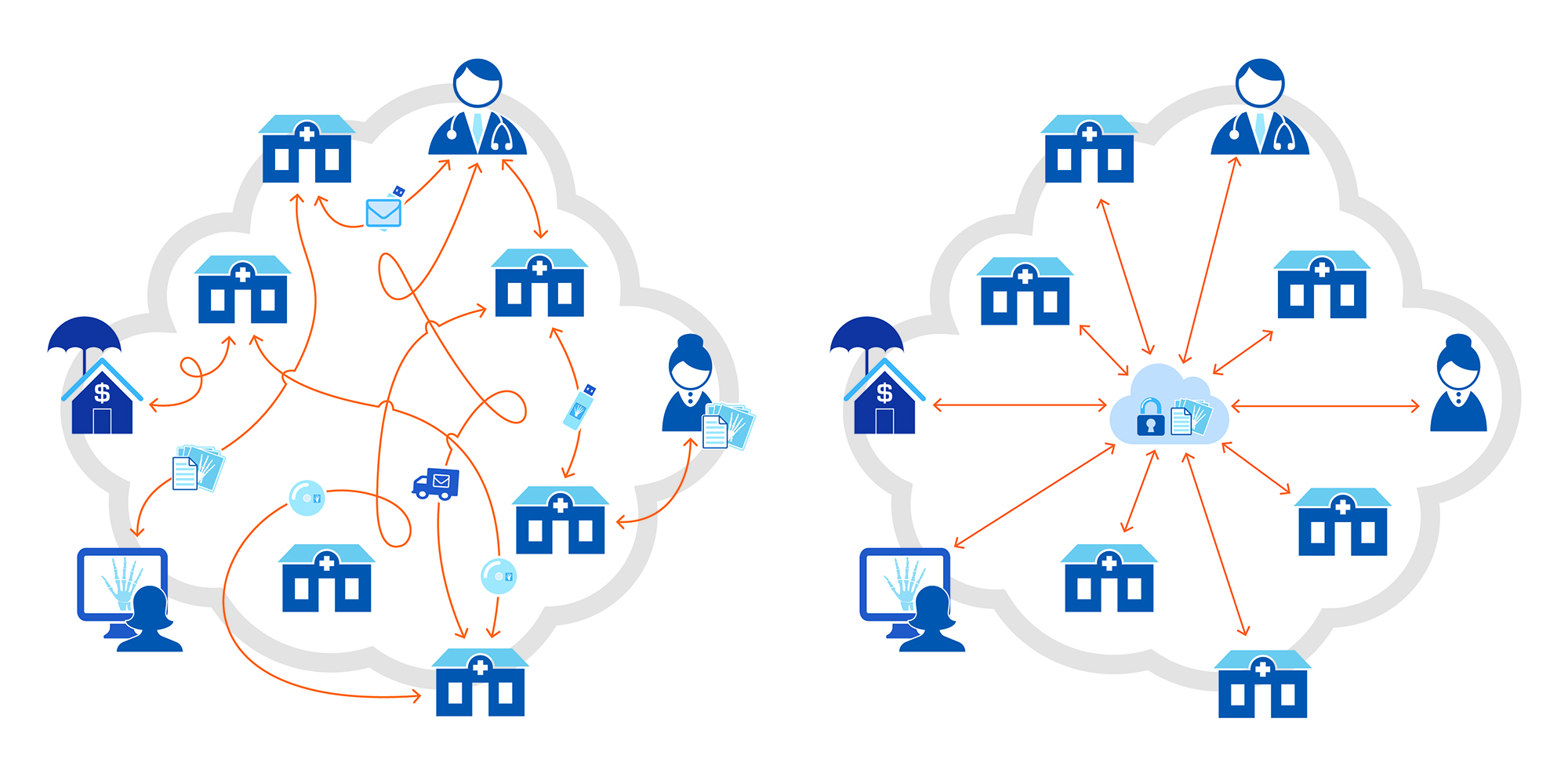
Commonly, our client-side logic will be found on the REPOSITORY_NAME/src/client/ folder. Inside this folder, we will have the css/ folder with all used stylesheets. Secondly, we will have the lib/ folder where we can find the client-side used libs. Thirdly, we will have a set of basic UI components. The set of UI components are fourfold: (1) the loaders/ folder; (2) the modals/ folder; (3) the tools/ folder; and (4) the viewport/ folder. Each folder represents a specific UI behavior described as follows. The loaders are the set of loading methods that will show the patient's lists consumed by the PACS, i.e., the DICOM servers. The modals are just windows pop-ups that we trigger when pressing expected links. The tools are the buttons and respective behavior of the buttons. Finally, the viewport is the set of files and methods where we manage the showing images.
Our prototypes support a complete imaging metric workflow. To streamline implementation, it was tested using various DICOM servers, such as the lightweight Orthanc server. By default, measurement data is stored in several JSON files database, which we call as datasets. Researchers can configure alternate data exchange mechanisms to support their datasets. Clinicians can leverage the prototype’s built-in datasets and DICOM servers to act as the central repository of DICOM images. Then any DICOM-compliant viewer or workstation can "pull" the datasets from the archive as needed. Accelerators may have significant constraints that limit the size of the image datasets they can render. Since loading images usually requires a call to a server, for the image loading we need to do it asynchronously. Our prototypes require that Image Loaders return an Object containing a Promise which the system will use to receive the Image Object asynchronously, or an Error if one has occurred. On the CornerstoneJS documentation, we find the same behavior workflow (see next Figure) followed by us.
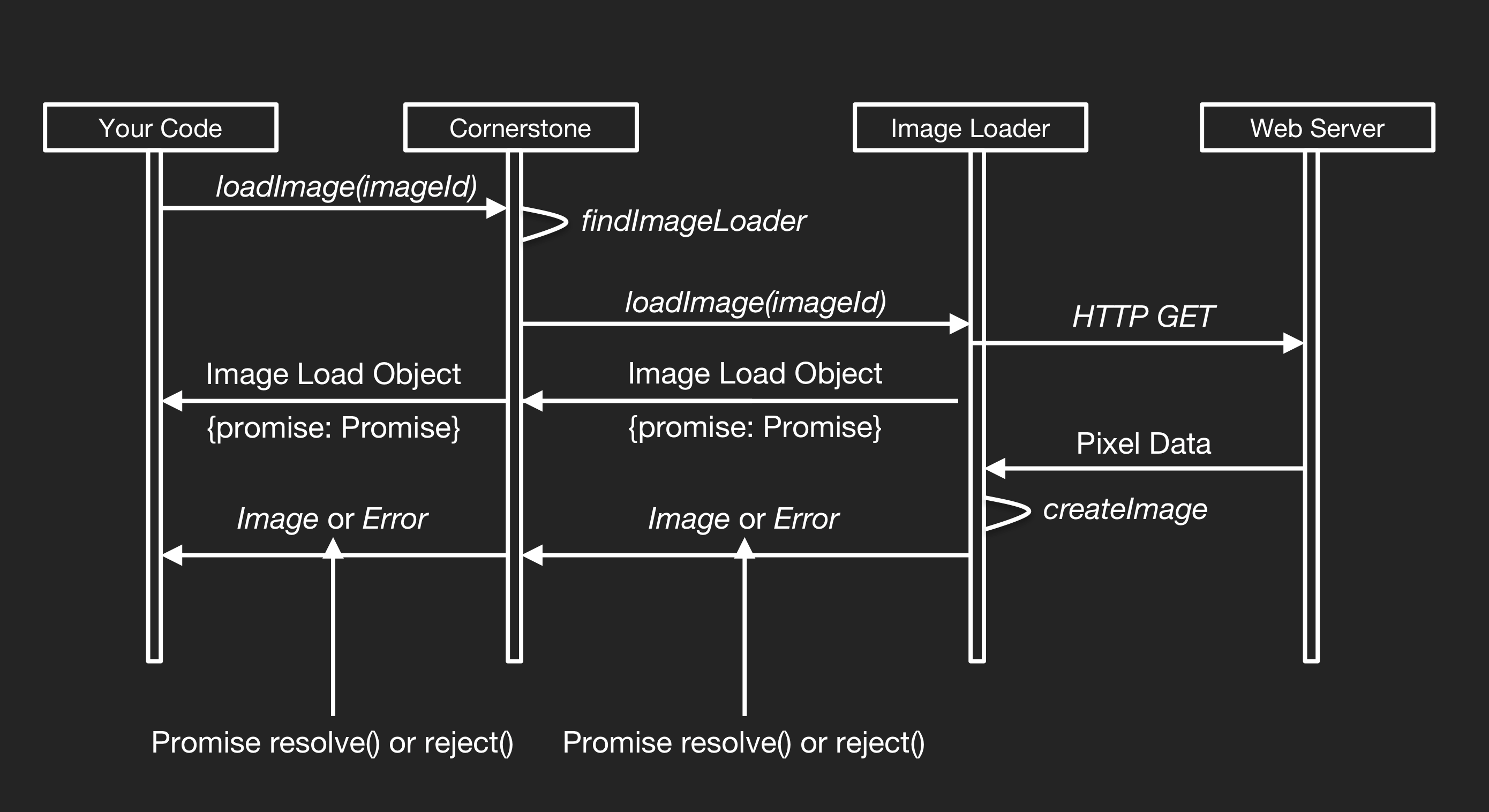
What we do is to register the ImageLoaders so that we can load specific ImageId URL schemes. In the end, the passed back Image Object will resolve the Promise and then display it using the displayImage() of the CornerstoneJS library. By default, the viewer is configured to a remote server connection. Each prototype runs the server application using NodeJS and can connect to DICOM Image Archives over both DICOM Service Exchange and DICOMWeb protocols. Nevertheless, we can find the full server-side logic on the REPOSITORY_NAME/src/server/ folder.