Welcome to the UCSF Data Science Initiative's single-cell RNA-sequencing workshop. This course is built from the following Satija Lab Seurat vignettes:
We add emphasis to areas of the vignettes we find most important, expand explanations in key areas, and add some of our own data processing and analysis if needed.
Team-reviewed, tested, and ready-to-use materials will always be hosted on our official UCSF Collaborative Learning Environment (CLE) scRNA-seq webpage.
This GitHub repository is for development purposes and therefore may stray from the materials used in class. CLE will remain as it was at the time of our most recently offered workshop and will only be updated if major changes are introduced to the course.
Branch "master" should function as intended if cloned, but for verified materials please download all documents and data from CLE unless otherwise instructed.
Please do not wait until the day of the workshop to do these installations! We cannot spend class time working through installation issues.
If you have trouble with installations up until the day before the workshop, please email the instructor:
- Karla Lindquist: karla.lindquist@ucsf.edu
Previous versions of this workshop relied on Python 3.x - Seurat has released an update to remove this dependency, so we no longer require a Python install of UMAP for this workshop.
We package R installations in .Rmd format - you'll need to have R and RStudio installed to proceed.
Open 0_workshop_prep.Rmd in Rstudio, run all the cells, and ensure all packages install without issue. Once done, you're ready for the workshop. And remember, please contact us ASAP if you have any trouble!
This workshop outlines the general workflow for analysis of scRNA seq data that that has already been preprocessed into either 10x Genomics' CellRanger output (.Rmd #s 1-5) or count matrices (.Rmd # 6).
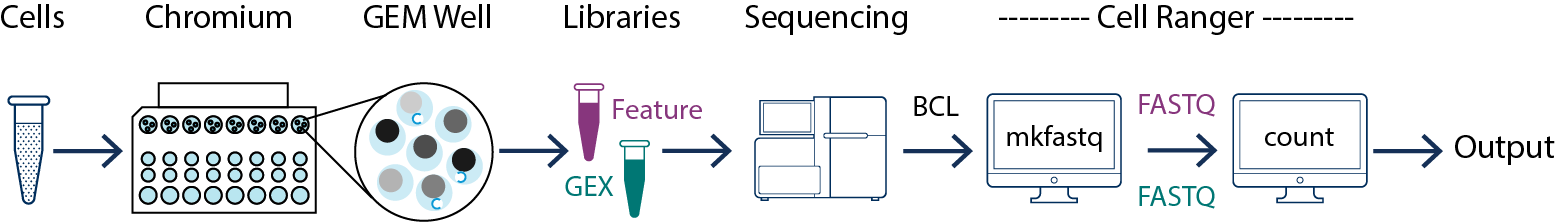
The majority of this workshop (notebooks 1-5) uses data output from the 10x Genomics CellRanger Pipeline. This software can demultiplexes, counts, and aggregates raw basecall data directly from Illumina sequencers:
Please note that this pipeline is not necessary for this workshop but will be needed to use your data with these notebooks. Visit the 10x Genomics CellRanger Pipeline website for installation instructions, user guides, and more information.
Seurat is an R toolkit for single cell genomics, developed and maintained by the Satija Lab at NYGC. Instructions, documentation, version history, and additional tutorials can be found on the Satija Lab Webpage. Seurat is also hosted on the Satija Lab GitHub, you can view and clone the repository here.
Seurat has been successfully installed on Mac OS X, Linux, and Windows, using the devtools package to install directly from GitHub. Improvements and new features will be added to the Satija Lab GitHub on a regular basis, please contact seuratpackage@gmail.com with any questions or if you would like to contribute.
Please be aware that GitHub is not certified for use with personal health information (PHI). Do not store or share any sensitive information via GitHub, even if kept in private repositories.
For more information regarding UCSF's security guidelines, please see the UCSF IRB guidelines for electronic data security.