You can create the docker image locally by either:
- (i) Building the docker image yourself via
docker build -t ads .Once you've got the docker image locally, you can run it via:docker run -w /ads -it --rm --gpus device=0 -v /local/path/to/ads:/ads/ -v /local/path/to/data/root/:/data/ ads:latest - (ii) Downloading the pre-built image from docker hub via
docker pull oisinnolan/ads. Similarly, you can run this via:docker run -w /ads -it --rm --gpus device=0 -v /local/path/to/ads:/ads/ -v /local/path/to/data/root/:/data/ oisinnolan/ads:latest.
This will spin up a docker container and open an interactive shell, within which you can run the scripts as usual using python.
The following sections explain how to prepare the fastMRI and MNIST datasets to reproduce the paper results. These datasets should be placed in some local directory, e.g. /data/ which we refer to as the 'data root'.
In order to create the fastMRI dataset used for the experiments in ADS, you'll need first to download fastMRI and then run the create_loupe_fastmri_dataset.py script in order to pre-process the data as was done in [1]. This will create a train/val/test split which can then be used for training the diffusion model and benchmarking against other methods.
- Download fastMRI at https://fastmri.med.nyu.edu/.
- Run
create_loupe_fastmri_dataset.py, replacingINPUT_ROOTwith the path to your local copy of the dataset, (the directory containingknee_singlecoil_train,knee_singlecoil_val, andknee_singlecoil_test), and desired output path.
- Download from http://yann.lecun.com/exdb/mnist/ and convert to png using, e.g. https://github.com/myleott/mnist_png, or download the png dataset directly via https://github.com/myleott/mnist_png/blob/master/mnist_png.tar.gz.
- By default the train and test sets are split into sub-directories by digit, but we flatten this structure into a single train directory and a single test directory via
create_mnist_dataset.sh.
- You can train your own diffusion model using
python train_ddim.py --config=/path/to/config --data_root=/path/to/data/root --run_dir=/path/to/save/model/. - For example, train on fastMRI using
train_ddim.py --config=configs/training/ddim_train_fastmri.yaml --data_root=data/ --run_dir=trained_models/. - Make sure to check that the
train_folderandval_folderin the config YAML file point to the correct datasets relative to your data root. e.g., if your data root is/data/, and thetrain_folderin your config isFastMRI/LOUPE/train, then the absolute path to your train set should be/data/FastMRI/LOUPE/train.
- Download link: https://drive.google.com/drive/folders/1URM_Agp-rQuEKlKkqopYYwGfkXp_pnYY?usp=drive_link
- Download the folder
trained_model, containing configs and weights for the MNIST and fastMRI models, and place this in the project root directory. The inference configs inconfigs/should then point to these directories.
- First, choose one of the configs in
configs/inferenceand make sure thatdiffusion_sampler.run_dirpoints to the folder containing your model'sconfig.yamland checkpoints directory. You can edit this config to change inference parameters, such as the number of samples to take, or number of reverse diffusion steps. - Then run inference using
python inference_active_sampler.py --config=/path/to/inference/config --data_root=/path/to/data/root --target_img=/path/to/target/img
e.g. python inference_active_sampler.py --config=configs/inference/mnist_pixels.yaml --data_root=data --target_img=sample_images/mnist_0.png
- Your results should be saved in your diffusion sampler run_dir directory, in the
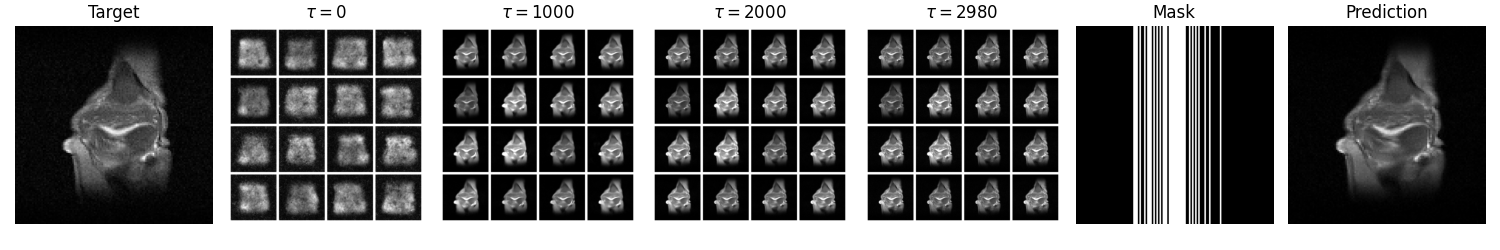
inferencesubdirectory. This should produce a number of outputs, including a summary GIF like the following:
- The two main experimental results from the paper are on (i) MNIST and (ii) fastMRI. These experiments have been implemented in the scripts
benchmark_mnist.pyandbenchmark_fastmri.py. There are then separate scripts that produce the plots and tables presented in the paper, from the benchmarking script outputs. - To reproduce the MNIST results, run:
# create mnist dataset variance map for fixed data-driven sampling
# NOTE: you must specify input and output directories in the file
python benchmark/create_mnist_dataset_variance_map.py
# to create results for each sampling strategy, run:
python benchmark/benchmark_mnist.py --sampling_interval=20 --test_set_path=data/MNIST/test --results_dir=results/MNIST --config=configs/benchmark/mnist/mnist_pixel_random.yaml
python benchmark/benchmark_mnist.py --sampling_interval=20 --test_set_path=data/MNIST/test --results_dir=results/MNIST --config=configs/benchmark/mnist/mnist_pixel_ads.yaml
python benchmark/benchmark_mnist.py --sampling_interval=20 --test_set_path=data/MNIST/test --results_dir=results/MNIST --config=configs/benchmark/mnist/mnist_column_random.yaml
python benchmark/benchmark_mnist.py --sampling_interval=20 --test_set_path=data/MNIST/test --results_dir=results/MNIST --config=configs/benchmark/mnist/mnist_column_ads.yaml
python benchmark/benchmark_mnist.py --sampling_interval=20 --test_set_path=data/MNIST/test --results_dir=results/MNIST --config=configs/benchmark/mnist/mnist_pixel_data_variance.yaml --dataset_variance_map_path=/path/to/data/variance.npy
python benchmark/benchmark_mnist.py --sampling_interval=20 --test_set_path=data/MNIST/test --results_dir=results/MNIST --config=configs/benchmark/mnist/mnist_column_data_variance.yaml --dataset_variance_map_path=/path/to/data/variance.npy
# Then, to produce the plots and table, run the following.
# Make sure to place all the .npy files output by benchmarking in the same results folder, and specify this in the file
# Also make sure to indicate whether the measurements are pixels or columns in MEASUREMENT_TYPE
python benchmark/make_mnist_results_plots.py- To reproduce fastMRI results, run:
# Run inference on the test set, save ssim results
python benchmark/benchmark_fastmri.py --use_test_set=True --test_set_path=data/FastMRI/LOUPE/test_hdf5/ --sampling_interval=1 --config=configs/benchmark/fastmri/fastmri_ads_10k.yaml --data_root=data/
python benchmark/benchmark_fastmri.py --use_test_set=True --test_set_path=data/FastMRI/LOUPE/test_hdf5/ --sampling_interval=1 --config=configs/benchmark/fastmri/fastmri_fixed_mask_10k.yaml --data_root=data/
# Aggregate SSIM results and create distribution plot
# Make sure to replace jobid with the jobid matching your benchmarking job
python benchmark/aggregate_fastmri_results.py --jobid=123456 --full_test_set=True --results_dir=results/fastMRI@article{nolan2024active,
title={Active diffusion subsampling},
author={Nolan, Oisin and Stevens, Tristan SW and van Nierop, Wessel L and van Sloun, Ruud JG},
journal={arXiv preprint arXiv:2406.14388},
year={2024}
}
[1] - End-to-End Sequential Sampling and Reconstruction for MR Imaging, Tianwei Yin*, Zihui Wu*, He Sun, Adrian V. Dalca, Yisong Yue, Katherine L. Bouman (*equal contributions). arXiv technical report (arXiv 2105.06460)