-
Notifications
You must be signed in to change notification settings - Fork 30
read analysis tutorial
For this tutorial, we'll begin with the same aligned, trimmed, and sorted BAM file, test.bam used in the Command Line Workflow tutorial, along with the corresponding SARS-Cov-2 reference genome.
Let's start off by finding covariant mutations (i.e. mutations present on the same sequenced read) on the spike gene. We can do so using the command:
freyja covariants freyja/data/test.bam 21563 25384 --output covariants_output.tsvThis saves covariance information to covariants_output.tsv, including the mutations present, their absolute count and frequency, as well as the genomic ranges covered by reads containing said mutations. To get a better sense of the data, we can generate a heatmap plot via:
freyja plot-covariants covariants_output.tsv --output covariants_heatmap.png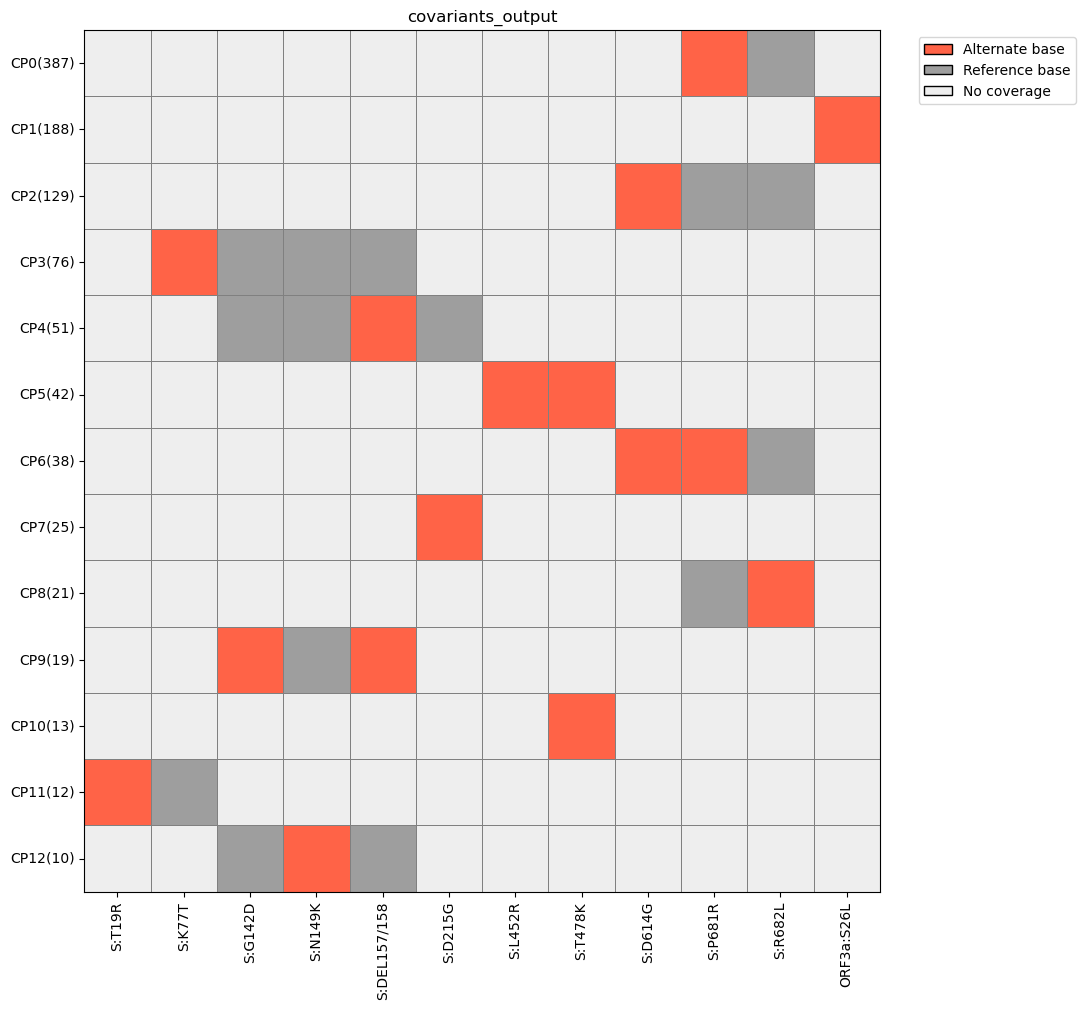
As we can see, there appear to be a number of mutations that occur together in this sample, such as S:D614G and S:P681R. We can pull these reads from test.bam using freyja extract. First, we must specify the mutations by creating a csv file, and then listing the corresponding nucleotide mutations, which are listed in covariants_output.tsv.:
A23403G,C23604G
We can now run freyja extract, passing in our query.csv file alongside test.bam. We'll include the --same_read flag since we're only interested in reads where both mutations are present
freyja extract query.csv freyja/data/test.bam --same_read --output extracted.bamFinally, we can view the pulled reads via IGV. First, we'll need to sort and index extracted.bam:
samtools sort extracted.bam > extracted_sorted.bam
samtools index extracted_sorted.bamNow load extracted_sorted.bam into IGV!
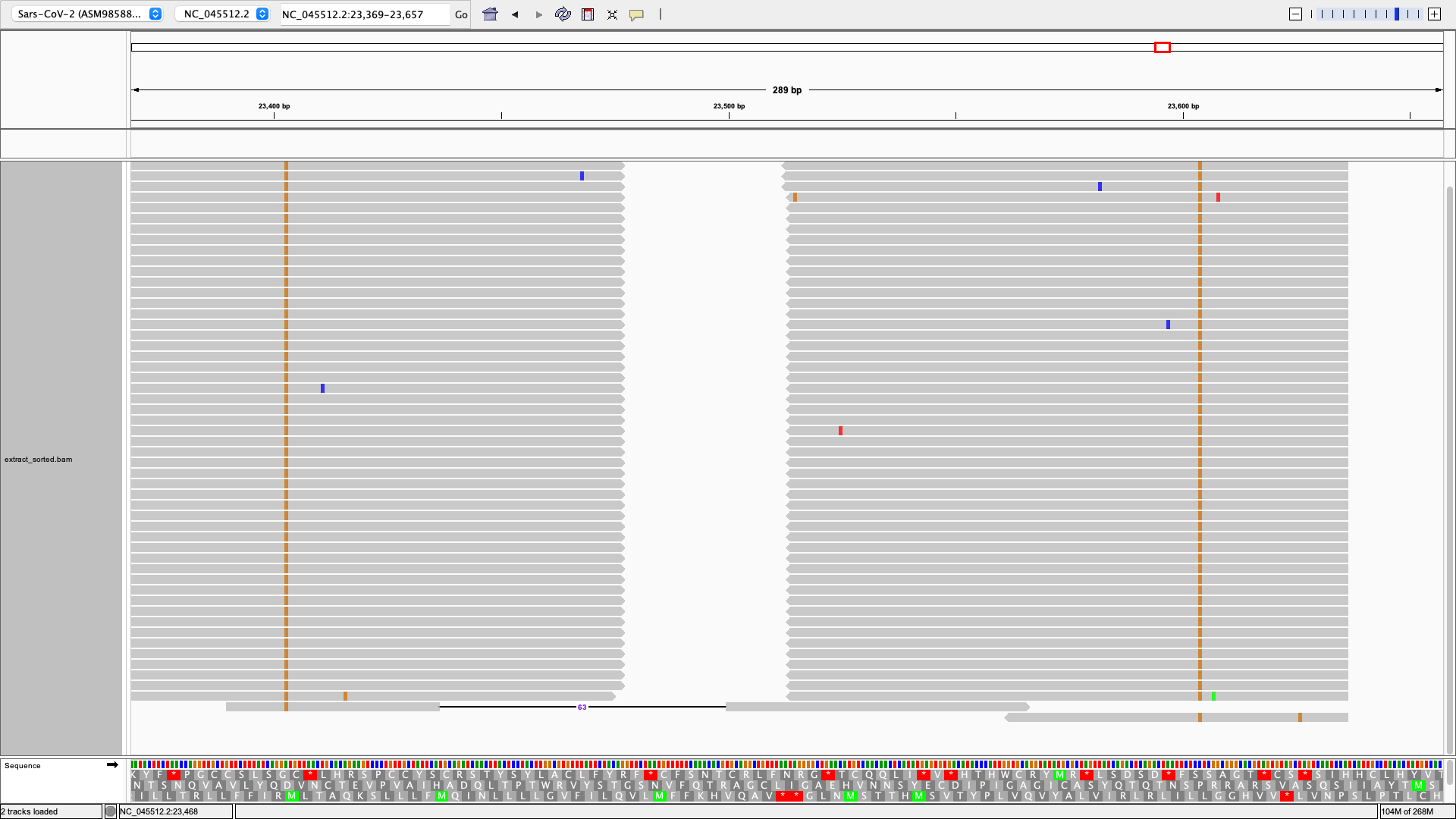
In some cases, it can be useful to remove reads containing one or more mutations. For instance, we might be interested in reads containing S:D614G but not S:P681R. We can do this by first calling freyja extract to select reads containing S:D614G, and then using freyja filter on the extraced output to filter out reads containing S:P681R.
- Extract S:D614G reads:
extract_query.csv:
A23403G
freyja extract extract_query.csv freyja/data/test.bam --output S_D614G_extracted.bam
samtools sort S_D614G_extracted.bam > S_D614G_extracted_sorted.bam
samtools index S_D614G_extracted_sorted.bam- Filter S:P681R reads from
S_D614G_extracted.bam
filter_query.csv:
C23604G
freyja filter filter_query.csv freyja/data/test.bam --output S_D614G_filtered.bamThe resulting S_D614G_filtered.bam should only contain reads containing S:D614G (which in this case happen to be unpaired reads with no coverage at S:P681).