-
Notifications
You must be signed in to change notification settings - Fork 2
Extensions
There are a number of extension packages that are linked in via the Extensions package. Most of these run via ProcessObject instead of LibraryLink simply because it was easier to write.
One of the external interfaces provided is to the electronic structure package Psi4. You can load the connection like this:
<<ChemTools`Extensions`Psi4`Install Psi4 if necessary:
Psi4Build[](*Out:*)
"Built to ~/Library/Mathematica/ApplicationData/ChemTools/Extensions/psi4/bin. Took 0 min."
If the system has to build and compile Psi4 you'll see a little window that tracks your progress:
There's a little wrapper for the Psi4 binary that actually calls Psi4
$Psi4(*Out:*)

This is just an alias for
Psi4@$Psi4Dir(*Out:*)

and you can supply any directory that provides access to bin/psi4 if you want to use a different Psi4 installation.
The Psi4 input file generator is a little bit complex. It uses the function Psi4ConfigureCall and a slew of options to set up an input file to be passed to Psi4. The way these options move from high- to low-level can be a bit involved.
The most bare-bones possible call provides a set of molecules (or single molecule) and a function to call.
Psi4ConfigureCall@
<|
"Molecules"->{{"O", {0, 0, 0}}},
"Function"-><|"Call"->"energy", "Arguments"->{"scf"}|>
|>(*Out:*)

If it is useful this can be elaborated upon at a later date. For the most part this has been handled and there will be a pre-made function to set up a run for whatever property is of interest.
We can then use the $Psi4 object to calculate things. Here's a way to run a single-point calculation on a molecule:
vcl = ImportString["vinyl chloride", "ChemObject"];
vclels = vcl["ElementPositions"];
out=$Psi4@Psi4Energy@vclels(*Out:*)

We can then access portions of this output. Here's a way to get the main output file data:
Short@out["output.dat"](*Out:*)
{<<1>>,<|"ZMatrix"->None,"MolTable"->None,<<1>>,"Properties"->{"M"…"p"->,<<14>>,<<1>>},"MetaInfo"->{}|>}
And we extract the energy from this:
(*Out:*)
-536.8736736700377605`18.729872108265432
And an energy of around -540 Hartrees doesn't seem ridiculous
Configure a dihedral scan in Psi4:
vclcom = vcl["CenterOfMass"];
scanels=
Append[
ChemTools`Formats`ChemFormatsConvert[Append[vclels, {"X", vclcom}], "ZMatrix"],
{"Ar", Length[vclels]+1, 3.1077, 2, 180, 1, #}
];
scan=
Psi4EnergyScan[
<|
"Molecules"->scanels,
"Scan"->{{0,180,60}},
"Configuration"->{
"BasisSet"->"cc-pvqz"
}
|>
](*Out:*)

Extract the input file string:
scan["input.dat"]//StringTrim-----------Out-----------
molecule mol {
Cl
C 1 1.71174
C 2 1.33244 1 2.15025
H 3 2.13278 2 0.43492 1 3.14148
H 4 2.48131 3 0.450246 2 3.1415
H 5 1.85827 4 1.56907 3 0.0000242934
X 6 2.13237 5 1.49914 4 0.000039543
Ar 7 3.1077 2 180 1 pyVar_1
}
set { basis cc-pvqz }
for ( pyVar_1 in range(0, 180, 60) ):
[ mol_1.pyVar_1 ] = [ pyVar_1 ]
energy('scf')The link provides access to the oeprop module in Psi4. These are the properties that may be expressed in terms of the one-electron wavefunctions.
out=$Psi4@Psi4OneElectronProperty[vclels, "Properties"->{"OrbitalExtents"}];
StringCases[
out["output.dat"],
which:DigitCharacter~~
Whitespace~~LetterCharacter~~Whitespace~~DigitCharacter~~
xyzr:Repeated[Whitespace~~NumberString, 4]:>
Floor@Internal`StringToDouble[which]->
AssociationThread[
{"x", "y", "z", "r"},
Map[Internal`StringToDouble, StringSplit[xyzr]]
]
](*Out:*)
{0-><|"x"->3.2466869184`,"y"->0.0621279459`,"z"->0.0037333295`,"r"->3.3125481939`|>,1-><|"x"->1.4312074702`,"y"->1.0532904221`,"z"->0.0325164356`,"r"->2.5170143279`|>,2-><|"x"->10.7134557718`,"y"->0.1915771908`,"z"->0.0325251933`,"r"->10.9375581558`|>,3-><|"x"->3.3110756455`,"y"->0.1350141301`,"z"->0.0771144181`,"r"->3.5232041937`|>,4-><|"x"->3.3607035999`,"y"->0.1124520668`,"z"->0.0408739769`,"r"->3.5140296436`|>,5-><|"x"->3.2836016075`,"y"->0.0991575718`,"z"->0.1223090113`,"r"->3.5050681906`|>,6-><|"x"->3.2963554217`,"y"->0.167966003`,"z"->0.0407649328`,"r"->3.5050863576`|>,7-><|"x"->3.0492260387`,"y"->1.1273477389`,"z"->0.8410559907`,"r"->5.0176297684`|>,8-><|"x"->7.9027010227`,"y"->1.2545937235`,"z"->0.7849140843`,"r"->9.9422088305`|>,9-><|"x"->7.8515070009`,"y"->3.8252578231`,"z"->0.7175615656`,"r"->12.3943263896`|>}
Access is provided to Psi4's geometry optimization routine:
out=$Psi4@Psi4GeometryOptimize[vclels];out["output.dat"][[-1, "MolTable"]](*Out:*)
{{"CL",{-0.987081306337`,0.138979445838`,-0.000029527506`}},{"C",{0.681519309539`,-0.55620763637`,0.000042522046`}},{"C",{1.764611233764`,0.202327834608`,0.000048927722`}},{"H",{0.665652931482`,-1.623935176841`,-0.000056319987`}},{"H",{2.73460576847`,-0.258132041658`,-0.000055614194`}},{"H",{1.723184459835`,1.273435633956`,0.000047583524`}}}
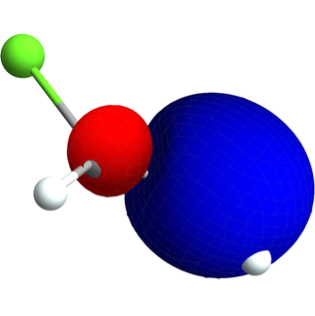
There are a number of high-level wrappers to Psi4 functionality provided, particularly via the object framework. Here's one use:
vcl["OrbitalsPlot"]["Orbitals"->{3}, "Mode"->"Cached"](*Out:*)
Along with the Psi4 connection there is a connection to OpenBabel , which delegates to the pybel Python module or the obabel CLI by defaut. You can load this like so:
<<ChemTools`Extensions`OpenBabel`If necessary, build and download OpenBabel
OBBuild[](*Out:*)
"Built to ~/Library/Mathematica/ApplicationData/ChemTools/Extensions/openbabel. Took 0.0000237 min."
As with psi4, when you build OpenBabel you'll see a little terminal window that tracks your progress:
As with the Psi4 object there's a custom OpenBabel object
$OpenBabel(*Out:*)

You can pass a custom one of these to the basic function OBRun function to use a different OpenBabel installation.
The core runners are the functions OBRun and OBPyRun , where OBPyRun simply layers on top of OBRun . The interface to OBRun is still a work in progress, so suggestions and clarifications are welcome.
Here's a way to convert a file from one format to another via the obabel CLI:
infile=
First@
URLDownload[
"https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/146261/record/SDF/?record_type=3d&response_type=save&response_basename=Structure3D_CID_146261",
FileNameJoin@{$TemporaryDirectory, "mox.sdf"}
];OBRun[infile, "OutputFile" -> "mox.pdb"](*Out:*)
"1 molecule converted"
FileNameJoin@{$TemporaryDirectory, "mox.pdb"}//ReadString//Snippet[#, 5]&-----------Out-----------
COMPND 146261
AUTHOR GENERATED BY OPEN BABEL 2.4.1
HETATM 1 O UNL 1 0.774 -0.756 -0.110 1.00 0.00 O
HETATM 2 C UNL 1 -0.212 0.063 0.503 1.00 0.00 C
HETATM 3 C UNL 1 0.973 0.663 -0.164 1.00 0.00 C Or get some basic properties via obprop :
OBRun[infile, "Mode"->"obprop"]~Snippet~5-----------Out-----------
name 146261
formula C3H6O
mol_weight 58.0791
exact_mass 58.0419
canonical_SMILES C[C@@H]1CO1 146261Or the molecule energy (from the MMFF94 force field) via obenergy :
OBRun[infile, "Mode"->"obenergy"]~Snippet~-8-----------Out-----------
TOTAL ANGLE BENDING ENERGY = 0.45338 kcal/mol
TOTAL STRETCH BENDING ENERGY = 0.00995 kcal/mol
TOTAL TORSIONAL ENERGY = 2.91510 kcal/mol
TOTAL OUT-OF-PLANE BENDING ENERGY = 0.00000 kcal/mol
TOTAL VAN DER WAALS ENERGY = 1.03092 kcal/mol
TOTAL ELECTROSTATIC ENERGY = 3.24469 kcal/mol
TOTAL ENERGY = 8.02257 kcal/molMore serious calculations can be done via the OBPyRun interface. It uses the PyTools package to run things through the pybel interface.
mox=ImportString["methyl-oxirane", {"ChemicalStructure", "ElementPositions"}];Here's an example of calculating the energy of methyl-oxirane off the UFF force-field:
OBPyRun[
mox,
ff = openbabel.OBForceField.FindForceField[String@"UFF"];
ff.Setup[pybelMol.OBMol];
ff.Energy[]//Print
](*Out:*)
1.31114*10^30
This is also one of the small number of routines cooked into the connection:
(*Out:*)
1.31114*10^30
In general OBPyRun delegates to RunProcess , but it's also possible to use a "Session" to speed things up. To do so we simply pass "Session"->True to any of the methods. If we do this a python runtime is kept alive in the background.
Here's an example:
OBPyRun[
mox,
ff = openbabel.OBForceField.FindForceField[String@"UFF"];
ff.Setup[pybelMol.OBMol];
ff.Energy[]//Print
]//RepeatedTiming(*Out:*)
{0.1337207777777777717`2.,1.31114137871`*^30}
OBPyRun[
mox,
ff = openbabel.OBForceField.FindForceField[String@"UFF"];
ff.Setup[pybelMol.OBMol];
ff.Energy[]//Print,
"Session"->True
]//RepeatedTiming(*Out:*)
{0.0264066315789473621`2.,1.31114137871`*^30}
And here's the runtime:
Processes[][[1]](*Out:*)
The session will keep track of any definitions we have made, e.g.:
OBPyRun[
mox,
ff = openbabel.OBForceField.FindForceField[String@"UFF"];,
"Session"->True
]Then if we check this definition later:
OBPyRun[
mox,
ff//Print,
"Session"->True
](*Out:*)
"<openbabel.OBForceField; proxy of <Swig Object of type 'OpenBabel::OBForceField *' at 0x1075f5c00> >"
We can also pass the key "Restart" to restart the session.
OBPyRun[
mox,
ff//Print,
"Session"->"Restart"
]OpenBabel::runerr: "Error in OpenBabel run:\n\n!(*RowBox[{"\"Traceback (most recent call last):\\n File \\\"\\\", line 2, in \\n File \\\"\\\", line 27, in \\nNameError: name 'ff' is not defined\""}])"
And if we pass the key "Close" it will remove the session:
OBPyRun[
mox,
ff//Print,
"Session"->"Close"
]Processes[](*Out:*)
{}