RABIES is an open source image processing pipeline for rodent fMRI. It conducts state-of-the-art preprocessing and confound correction, and supplies standard resting-state functional connectivity analyses. Visit our documentation at https://rabies.readthedocs.io/en/stable/.
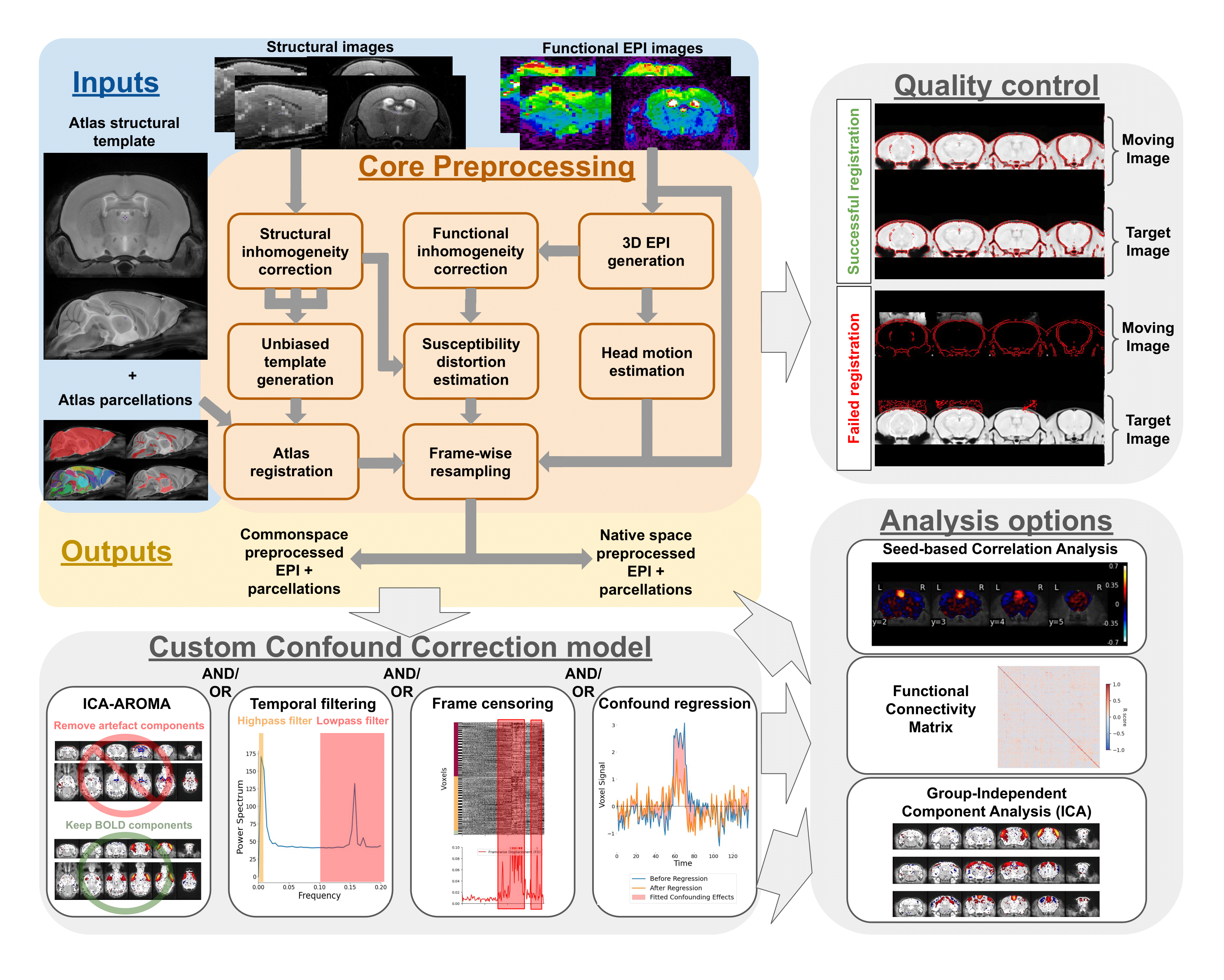
The primary purpose of RABIES is to provide rodent fMRI research with a standard, flexible, and reliable image processing platform. The package is complemented with informative data diagnostic features for confound characterization and encourages best practices for quality control and reproducibility. The RABIES software is structured into three main processing stages: preprocessing, confound correction and analysis.
The preprocessing workflow regroups essential fMRI preprocessing steps prior to analysis. It includes a robust registration workflow with automatically-adapting parameters allowing to succesfully process diverse acquisition types (i.e. rodent species, scan field strength, coil type, ...), and can conduct the following preprocessing steps:
- head motion correction
- susceptibility distortion correction
- resampling to native or common space
- brain parcellation
- slice timing correction (optional)
- despiking (optional)
- visual assessment of registration for quality control
Following preprocessing, a range of strategies to correct fMRI confounds (e.g. motion) can then be conducted within RABIES:
- linear detrending
- confound regression (with several options for nuisance regressors)
- frequency filtering (highpass, lowpass, bandpass)
- frame censoring (or scrubbing)
- ICA-AROMA
- spatial smoothing
Simple resting-state connectivity analyses are made available after preprocessing and confound correction. RABIES also provides a 'data diagnosis' workflow, which generates several indices of data quality and potential confounds, and conversaly, aims to improve the correction of confounds and transparency with regards to data quality:
- seed-based functional connectivity
- whole-brain connectivity matrix
- group-ICA
- dual regression
- data diagnosis
Nipype workflows: The image processing pipelines are structured using the Nipype library, which allows to build dynamic workflows in the form of a computational graph. Each node in the graph consists of a processing step, and the required inputs/outputs define the links between nodes. In addition to supporting code organization, Nipype workflows also handle several plugin architectures for parallel execution as well as memory management. The computational time to run the entire RABIES pipeline will vary substantially depending on data size, but for most uses, it will range from a few hours to a day when using proper computational resources and parallel execution.
Reproducible and transparent research: RABIES aims to follow best practices for reproducible and transparent research, including the following:
- open source code https://github.com/CoBrALab/RABIES
- standardized input data format with BIDS
- easily shared, automatically-generated visual outputs for quality control
- containerized distribution of the software hosted on Docker Hub which can be downloaded via Docker and Singularity platforms
Citing RABIES: A preprint introducing the software is available on bioXriv, which should be cited accordingly when using RABIES.
Boilerplate: a boilerplate summarizing the preprocessing and confound correction operations is automatically generated in the output folder. You can use the boilerplate to help describe your methods in a paper.
The RABIES license allows for uses in academic and educational environments only. Commercial use requires a commercial license from CoBrALab contact@cobralab.ca, http://cobralab.ca
This software was developped by the CoBrALab, located at the Cerebral Imaging Center of the Douglas Mental Health University Institute, Montreal, Canada, in affiliation with McGill University, Montreal, Canada. This work was supported by funding from Healthy Brains, Healthy Lives (HBHL), the Fonds de recherche du Québec - Santé (FRQS) and - Nature et technologies (FRQNT), and the Natural Sciences and Engineering Research Council (NSERC) of Canada. fMRIPrep was an important inspirational source for this project, in particular with regards to best practices for software reproducibility and code design using Nipype. We also thank the organizers of BrainHack School Montreal, which guided the initial steps of this project in 2018.
If you need support in using the software or experience issues that are not documented, we'll provide support on the Github discussion.
RABIES is under continuous development to keep up with evolving demand and ongoing research. We welcome suggestions for improvements using the Github issue system. If you're interested in contributing code, you can reach out on the Github discussion and we welcome contributions as pull requests.