Project made by Beatriz Soviero and Lais Canabarro
- Description of the RNA Translation Process
- Deterministic Finite Automaton
- Automaton Implemented with Python
- How to Use the Program
RNA its a molecule that synthesizes proteins. Its structure is formed by a single strand that contains four types of nitrogenous bases:
- Adenine (A)
- Cytosine (C)
- Guanine (G)
- Uracil (U)
To synthesize proteins, this process needs two phases:
- Transcription - synthesis of messenger RNA (mRNA) by reading DNA´s nitrogenous bases
- Translation - the chain of bases contained in the mRNA are divided in groups of 3 bases (called codons) that are translated into amino acids. This stage begins after reading the start codon (AUG) and ends after reading a stop codon (UAA, UAG or UGA). The protein is a result of the group of amino acids encoded by the read codons.
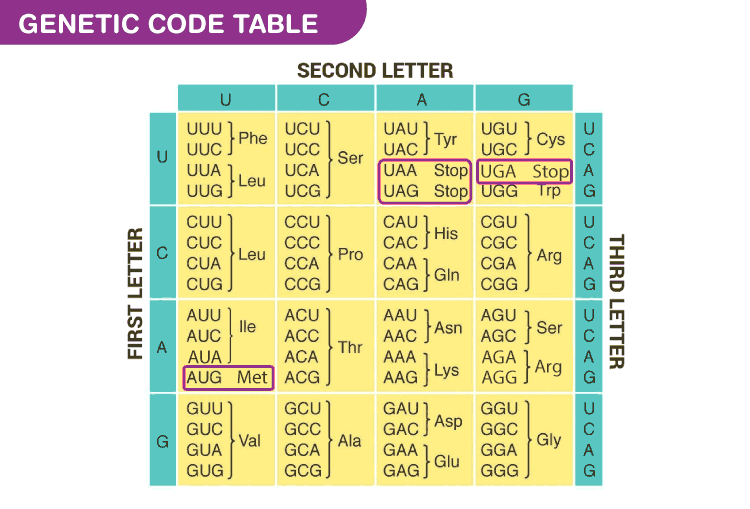
The relations among codons and amino acids are described in the genetic code, shown in the table below:
The group of symbols accepted by our automaton are the four types of nitrogenous bases (A, U, C, G) and all the acaminoacids present in the genetic code (Phe, Leu, Ser, Tyr, Cys, Trp, Pro, His, Gln, Arg, Ile, Thr, Asn, Lys, Val, Ala, Asp, Glu, Gly)
The automaton´s start stage is the q0 (when the start codon is read) and the final states q28 and q30 (when a stop codon is read)
The class Automato is initialized with the parameters:
- estados (array of strings that represent states);
- alfabeto (array of strings that represent accepted symbols);
- q0 (string with the initial state);
- fim (array of strings that represent final states);
- transicoes (dictionary in the format {('state', 'symbol') : 'next state'}).
class Automato:
def __init__(self, estados, alfabeto, q0, fim, transicoes):
self.estados = estados
self.alfabeto = alfabeto
self.q0 = q0
self.fim = fim
self.transicoes = transicoes
self.atual = q0
In addition, Automato also contains flags that represent the acceptance or not in the format [boolean, "message of acceptance or error"]:
self.ACEITA = [False, 'ACEITA!']
# Check if empty word is accepted
if q0 in fim:
self.ACEITA[0] = True
# Cases when the word is rejected
self.NAO_FINAL = [not(self.ACEITA), 'REJEITA! Atingiu estado não final! O códon de parada não foi lido']
self.INDEFINIDO = [False, lambda s, c: f'REJEITA! O aminoácido {s} não é codificado pelo códon {c}']
self.ALFABETO = [False, lambda s: f'REJEITA! O símbolo {s} não está contido no alfabeto']
self.ERRADO = False
To execute our automaton, we created the step function that evaluates the given symbol and, if accepted, goes to the next state.
# sigla and codon are used when an amino acid is read. In this case sigla=True and codon= codon that encodes it
def step(self, simbolo, sigla = False, codon = None):
if simbolo not in alfabeto: # When symbol is not in the alphabet
self.ALFABETO[0] = True
print(self.ALFABETO[1](simbolo))
self.ERRADO = True
return
#proxEstado = None means undefined transition
try:
proxEstado = self.transicoes[(self.atual, simbolo)]
except KeyError:
proxEstado = None
# When the next state doesn´t exist
if proxEstado is None:
self.INDEFINIDO[0] = True
if sigla: print(self.INDEFINIDO[1](simbolo, codon))
else: print('REJEITA! Transição indefinida')
self.ERRADO = True
return
#If next state is not final, keeps reading
if proxEstado not in self.fim:
self.NAO_FINAL[0] = True
#If next state is finall, input is accepted
if proxEstado in self.fim:
self.ACEITA[0] = True
return
if sigla:
print(f'Códon {codon} e aminoácido {simbolo}: OK')
self.atual = proxEstado
There are two ways for reading the RNA chain:
- Terminal reading (1): to get the input by the terminal, there are five steps to be followed
- Type the start codon;
- In the next line, write a codon and press ENTER;
- In the next line, the amino acid encoded by step 2 and press ENTER;
- Repeat steps 2 and 3 as many times as necessary;
- Type the stop codon.
- File reading (2): to read a chain written in a file, .csv and .txt formats are supported. You only need to type the file´s path.