This REANA reproducible analysis example emulates a typical Beyond Standard Model (BSM) search as performed in collider particle physics. It involves processing three main groups of data:
- The observed collision data as it was recorded from the detector
- The Standard Model Backgrounds relevant for the search
- The Beyond Standard Model signal sample.
After processing, a statistical model involving both signal and control regions is built and the model is fitted against the observed data. In this emulation, the data is compatible with the Standard Model expectation, and thus an upper limit on the signal strength of the BSM process is computed, which is the main output of the workflow.
This example uses the ROOT data analysis framework and Yadage computational workflow engine.
Making a research data analysis reproducible basically means to provide "runnable recipes" addressing (1) where is the input data, (2) what software was used to analyse the data, (3) which computing environments were used to run the software and (4) which computational workflow steps were taken to run the analysis. This will permit to instantiate the analysis on the computational cloud and run the analysis to obtain (5) output results.
In this example, the input datasets representing the collision and simulated data will be generated on the fly in the first couple of workflow steps. Therefore there is no explicit input data to be taken care of.
This example uses the ROOT analysis framework with the custom
user code located in the code directory. In order to execute the different stages of
the analysis a number of scripts are needed. In a real analysis these scripts might be
part of larger analysis frameworks developed using the experiment-internal software stack
(e.g. CMSSW or the ATLAS Analysis Releases) and be based on C++ with many dependencies
and require multiple container images. In this emulation we have two container images.
- A pure ROOT6 container image
docker.io/reanahub/reana-demo-bsm-searchused for most steps (such as selection, merging etc) - An image based on ROOT6 which also has the
hftoolspackage installed. This image is used for the last steps dealing with fitting, plotting and exporting to HepData
generantuple.py - Generating Toy Data
This script generates toy datasets needed for the analysis. The script has the command line interface
python /code/generantuple.py {type} {nevents} {outputfile},
where {type} can be one of [data, mc1, mc2, qcd, sig] generating "observed data", two
background processes "mc1" or "mc2", a "multijet-background" and finally the BSM signal
process, respectively. The "data" is just a specific mix of the other three processes
according to their respective cross sections.
The dataset which is a collection of "events" (the number of events is controlled by the
{nevents} parameters) which is stored in a ROOT TNtuple at the path indicated by
{outputfilename}.
Since dataset generation is easily parallelizable, ultimately we will run many of these
jobs at the same time and merge the TNtuples via ROOT's hadd utility.
select.py - Selecting Interesting Events
This is the main "event selection" code that processes the datasets and selects "interesting events" and applies correction and systematic variations to the events. In a real analysis this would be the bulk of the analysis code implemented in a C++ experiment framework. In this example, the cli structure of the script is
python /code/select.py {inputfile} {outputfile} {region} var1,var2,...
where an input and output files are specified as well as the region (i.e. either signal or control region) and a number of comma-delimited systematic variations are specified. The code then applies cuts and variations to the events and writes the selected events into a new TNtuple which is saved to disk. In this case only variations that affect the event selection need to be specified. Variations that only affect the event weights are dealt with in the histogramming step (see below).
histogram.py - Summarize Events in histograms
This script reads in the TNtuple of the selected events and creates the required histograms for building the statistical model and weights them to a specific luminosity. The command structure is
python /code/histogram.py {inputfile} {outputfile} {name} {weight} var,var2,...
the variations in this case are weight-only variations.
makews.py - Building a Statistical Model
This script creates a RooWorkspace using the HistFactory p.d.f template. The
HistFactory configuration has a single channel and four samples (qcd, mc1, mc2 and
signal). The parameter of interest in this model is the normalization of the signal
sample (the signal strength). For fitting and plotting the resulting workspace we use an
external package called hftools (HistFactory tools), which provides command line tools
for these purposes and no additional code is needed from our side. The command line
structure is
python /code/makews.py {data_bkg_hists} {workspace_prefix} {xml_dir}
The script expects all data and background histograms to be collected in a single ROOT file and writes the XML configuration and workspace to the paths specified on the command line.
hepdata_export.py - Preparing a HepData submission
The final step of an analysis is often to prepare a HepData submission in order to
archive measured distributions and results on the HepData archive. Here we use hftools
as a python library in this script, which has some convenience functions to generated
HepData tables from a RooWorkspace.
The command line structure is:
python /code/hepdata_export.py {combined_model}
In order to be able to rerun the analysis even several years in the future, we need to "encapsulate the current compute environment", for example to freeze the ROOT version our analysis is using. We shall achieve this by preparing a Docker container image for our analysis steps.
Some of the analysis steps will run in a pure ROOT analysis environment. We can use an already existing container image, for example reana-env-root6, for these steps.
Some of the other analysis tasks wil need hftools Python library installed that our
Python code needs. We can extend the reana-env-root6 image to install hftools and to
include our own Python code. This can be achieved as follows:
$ less environments/reana-demo-bsm-search/Dockerfile# Start from the ROOT6 base image:
FROM docker.io/reanahub/reana-env-root6:6.18.04
# Install HFtools and its dependencies:
RUN apt-get -y update && \
apt-get -y install \
libyaml-dev \
python-numpy \
zip && \
apt-get autoremove -y && \
apt-get clean -y
RUN pip install hftools==0.0.6
# Mount our code:
ADD code /code
WORKDIR /codeWe can build our analysis environment image and give it a name
docker.io/reanahub/reana-demo-bsm-search:
$ docker build -f environment/Dockerfile -t docker.io/reanahub/reana-demo-bsm-search .We can push the image to the DockerHub image registry:
$ docker push docker.io/reanahub/reana-demo-bsm-search(Note that typically you would use your own username such as johndoe in place of
reanahub.)
This analysis example intends to emulate fully what is happening in a typical BSM search analysis. This means a lot of computational steps with parallel execution and merging of results.
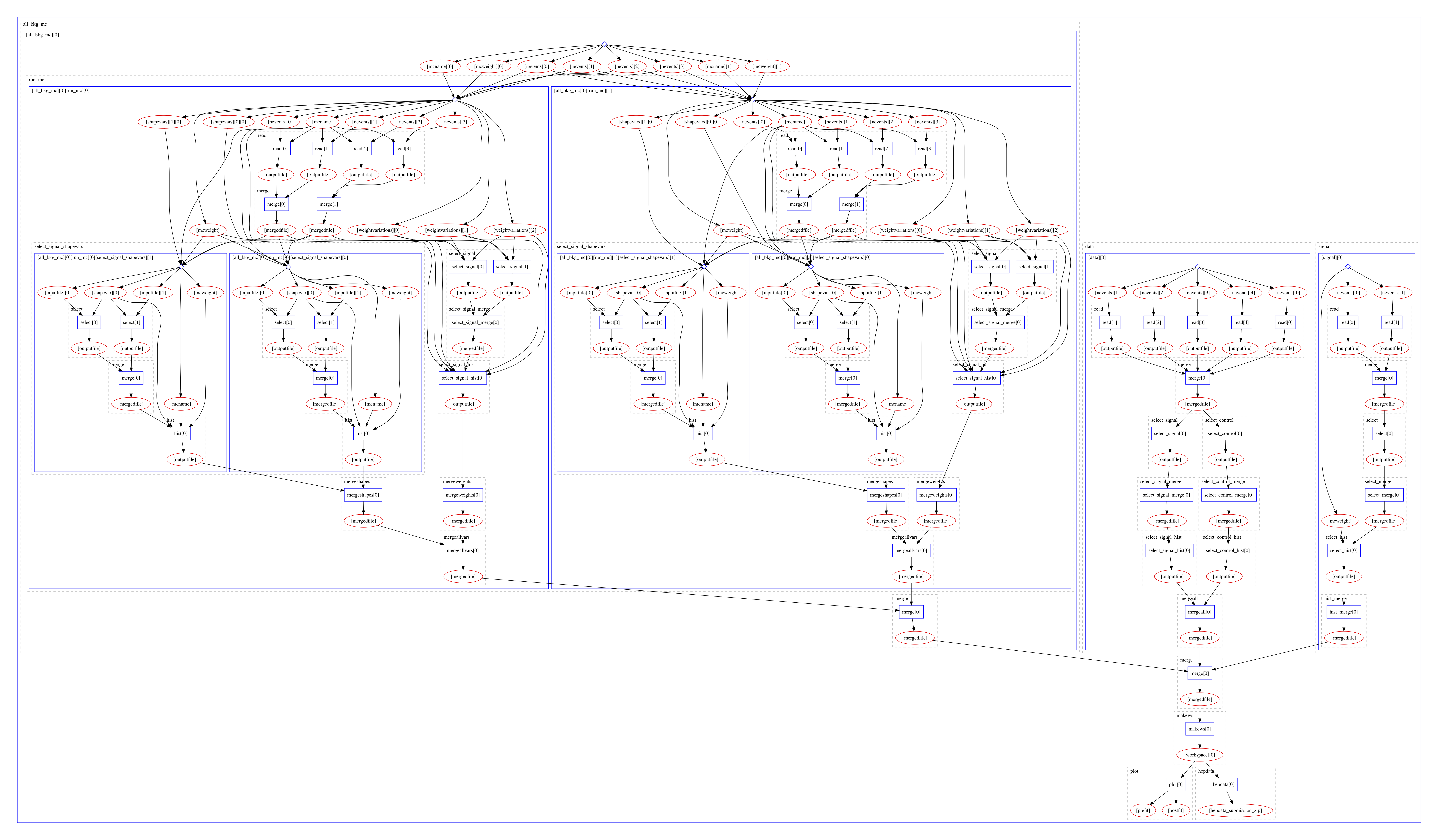
We shall use the Yadage workflow engine to express the computational steps in a declarative manner. The databkgmc.yml workflow defines the full pipeline defining various data, signal, simulation, merging, fitting and plotting steps:
At a very high level the workflow is as follows
-
Generate and process "observed data" to produce observed data and a data-driven multijet estimate in the signal region.
-
For each non-multijet Standard Model process (MC1 and MC2), generate and process datasets including systematic variations
-
Generate and Process a signal dataset
The three sub-workflows above can happen in parallel as they are independent of each other. Once they are done the remaining steps needed are
-
Merge Outputs from subworkflows and prepare a Statisical Model.
-
Perform Fits and produce Plots.
-
Prepare a HepData Submission
+---------------+ +--------------+ +------+
| Data & Mulijet| |SM Backgrounds| |Signal|
+---------------+ +--------------+ +------+
| | |
| | |
+--------> v <----------+
+--+--+
|Merge|
+--+--+
|
v
+----------+
| Workspace|
+----------+
+-----------+ | | +------------------+
|Fit & Plots| <---+ +----> |HepData Submission|
+-----------+ +------------------+The subworkflow generating and processing the "observed data" goes through these high-level stages.
- Generating the Data This stage generates data in a highly parallel fashion and then merges the files into a smaller number of files. We do not merge into a single file as this may end up being too large (currently merges happen in batches of six)
- Processing Data in Signal Region This branch in the data workflow processes the data and selects and histograms events in the signal region. This will be the data the model is fitted against.
- Processing Data in Control Region for data-driven multijet estimate This branch selects and histograms events in the control region to estimate the shape of the distribution and then uses a transfer factor which controls the normalization of the distribution in the signal region. This results in a so-called "data-driven" estimate the so-called "multijets" (or "qcd") background, since it would be unfeasible to estimate it using Monte-Carlo samples.
- Merge final results Finally, the results are merged into a single file that holds all the resulting histograms from the data sub-workflow.
For each of the SM backgrounds that are not estimated directly from the data, we use generated Monte-Carlo samples. For the Standard Model backgrounds we generate and process these datasets including systematics variations. These systematic variations change the values of the variables that are used to select "interesting events" as well as the "weight" of the event that is used when filling the histograms.
The SM Background sub-workflow splits into further sub-sub-workflows performed for each of the background processes. In this emulation we have two such processes.
For each sample, we go through the following stages
- Generate datasets for the background processes
- Run Event selection for Signal region
- Histogram Events (with correct luminosity weighting)
As some systematics affect the variables that are cut on in the event selection ( so-called shape variatiosn), the event selection step needs to be performed multiple times (once for each shape variations). Therefore, there is an additional sub-workflow for processing shape variations.
Systematics only affecting the weights can be implemented in one go at the histogramming stage.
As we progress through these stages, we add merging steps to reduce the number of files that need to be handled.
Finally, all histograms for a single Monte Carlo samples are collected before merging all Monte-Carlo samples into a single ROOT file.
The Signal workflow is very similar to the SM Background workflow, but we do not consider any systematics. Therefore it is a simple workflow that selects and histograms events (with a couple of merge stages in between).
Using these sub-workflows, we assemble a composed workflow. In this example, there are no externally settable parameters, as the parameters for the three sub-workflows (data, backgrounds, signal) are fixed in the workflow spec.
The parameters for the subworkflows include information on how many events to generate and, in the case of signal and background, what the relative weight should be.
$ head -8 workflow/databkgmc.yml
stages:
- name: all_bkg_mc
scheduler:
scheduler_type: singlestep-stage
parameters:
mcname: [mc1,mc2]
mcweight: [0.01875,0.0125] # [Ndata / Ngen * 0.2 * 0.15, Ndata / Ngen * 0.2 * 0.1] = [10/16*0.03, 1/16 * 0.02]
nevents: [40000,40000,40000,40000] #160k events / mc samplePlease see the databkgmc.yml workflow definition and related Yadage documentation.
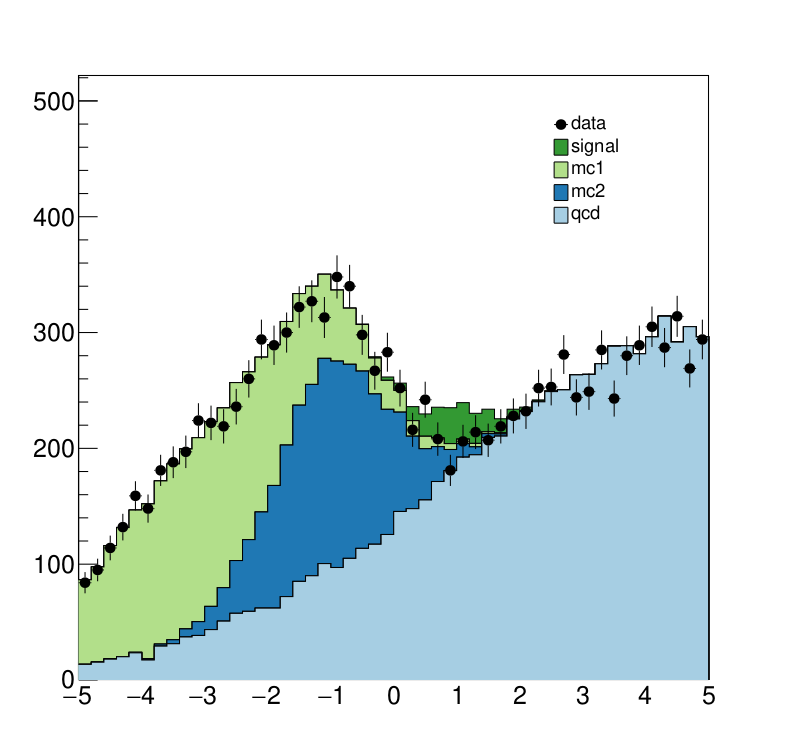
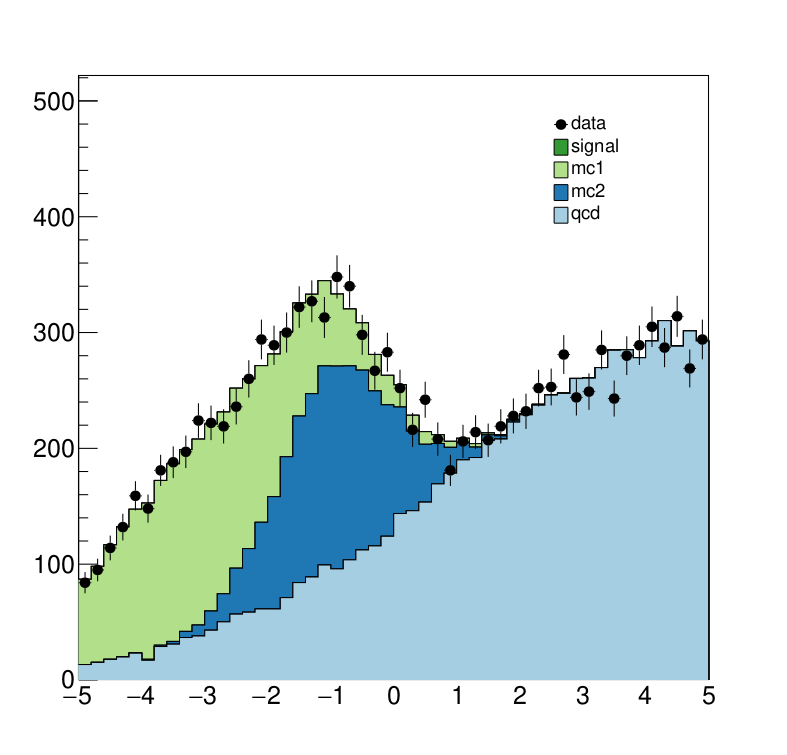
The interesting fragements generated by this result are the pre- and the post-fit distributions of the individual samples as well as the HepData submission in the form of a ZIP archive.
Below we see the model at its pre-fit configuration at nominal signal strength mu=1. The signal distribution is shown in green. As we can see the nominal setting does not describe the data, which is shown in black dots, well.
Here we see the post-fit distribution. As we can see, the signal sample needed to be scale down significantly to fit the data, which is expected since we generated the data in accordance with a SM-only scenario.
There are two ways to execute this analysis example on REANA.
If you would like to simply launch this analysis example on the REANA instance at CERN and inspect its results using the web interface, please click on the following badge:
If you would like a step-by-step guide on how to use the REANA command-line client to launch this analysis example, please read on.
We start by creating a reana.yaml file describing the above analysis structure with its inputs, code, runtime environment, computational workflow steps and expected outputs:
version: 0.6.0
inputs:
directories:
- workflow
workflow:
type: yadage
file: workflow/databkgmc.yml
outputs:
files:
- plot/prefit.pdf
- plot/postfit.pdfWe can now install the REANA command-line client, run the analysis and download the resulting plots:
$ # create new virtual environment
$ virtualenv ~/.virtualenvs/myreana
$ source ~/.virtualenvs/myreana/bin/activate
$ # install REANA client
$ pip install reana-client
$ # connect to some REANA cloud instance
$ export REANA_SERVER_URL=https://reana.cern.ch/
$ export REANA_ACCESS_TOKEN=XXXXXXX
$ # create new workflow
$ reana-client create -n my-analysis
$ export REANA_WORKON=my-analysis
$ # upload input code and data to the workspace
$ reana-client upload
$ # start computational workflow
$ reana-client start
$ # ... should be finished in about 15 minutes
$ # check its status
$ reana-client status
$ # list workspace files
$ reana-client ls
$ # download generated plots
$ reana-client downloadPlease see the REANA-Client documentation for
more detailed explanation of typical reana-client usage scenarios.