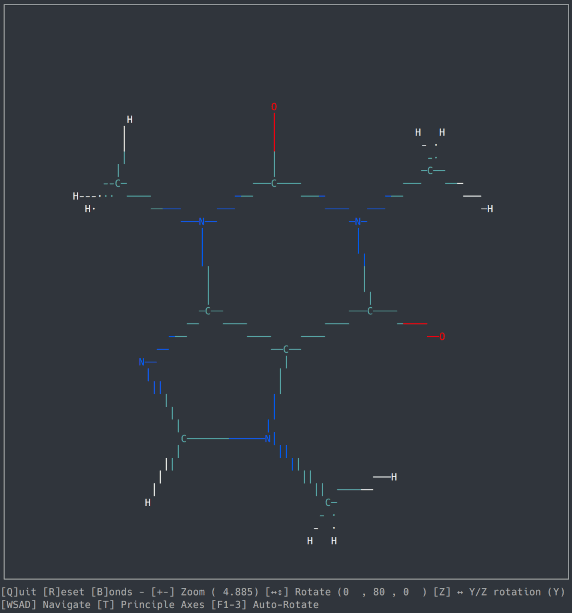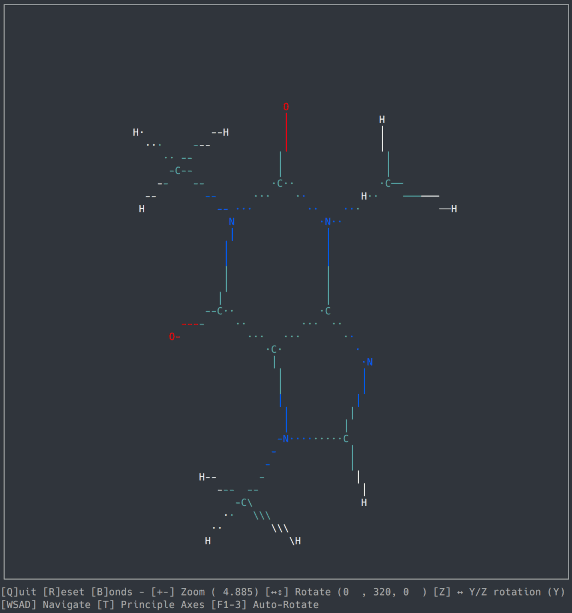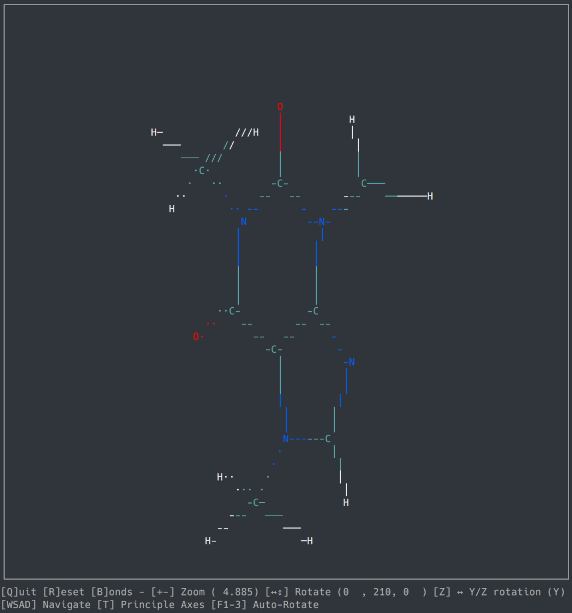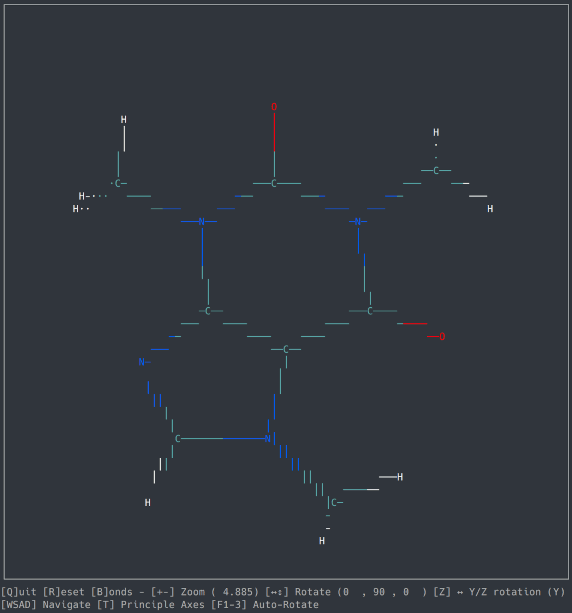
A basic molecule viewer written in Python, using the ncurses library. Works on any compatible terminal (even on Windows using windows-curses).
Features:
- Opening default cartesian .xyz files
- Orthographic view
- Navigation
- Zoom, Rotation, Auto-Rotation
- Bond detection and display
- Support for simple .xyz trajectories
- Optional integration of ASE and RDKit pypi packages for more formats and SMILES
pip install asciimol(Note: pip will install a run script in $HOME/.local/bin/ if you do not install with root permissions, so make sure this directory is part of your $PATH.)
You can also run
pip install asciimol[formats,smiles]to automatically install ASE for formats and RDKit for smiles.