AMPml AMPml is a software for antimicrobial peptide (AMP) predicting written in python.
AMPml operates in the Python environment, and run on the Windows system.
Step 1. Enter the command in CMD to open the visual interface: python main.py
Step 2. Choose prediction model (PAAC or CTDD).
Step 3. Choose input sequences file (and drop features file).
AMPml integrates four machine-learning algorithms (including random forest, support vector machines, gradient ascent and naive bayes classifier) and three amino acid feature descriptors (including AAC (20), PAAC (20+lambda) and CTDD (175)).
- Two public AMP databases APD3 and dbAMP were merged as a big AMP dataset.
- The sequences length less than 10 or longer than 100 were filtered out.
- The sequences with “X” (any amino acid) were deleted.
- The redundant sequences were filtered out.
- The protein dataset was downloaded from the public database Uniprot (100).
- The protein dataset was filtered by a R package ampir (https://github.com/Legana/AMP_pub/blob/master/02_build_training_data.md).
- The sequences length less than 10 or longer than 200 were filtered out.
- The sequences with “X” (any amino acid) were deleted.
steps 1-4 were the same as “Negative training dataset 1”. 5. Start codon “M” were deleted from each protein sequence.
The length of two training datasets
- According to consult literatures and practice, Positive training dataset/ Negative training dataset =1:3, the accuracy of PAAC is the highest; Positive training dataset/ Negative training dataset =1:1, the accuracy of CTDD is the highest.
- Random forest always achieved the best performance.
- The effect of amino acid feature descriptors was not significant.
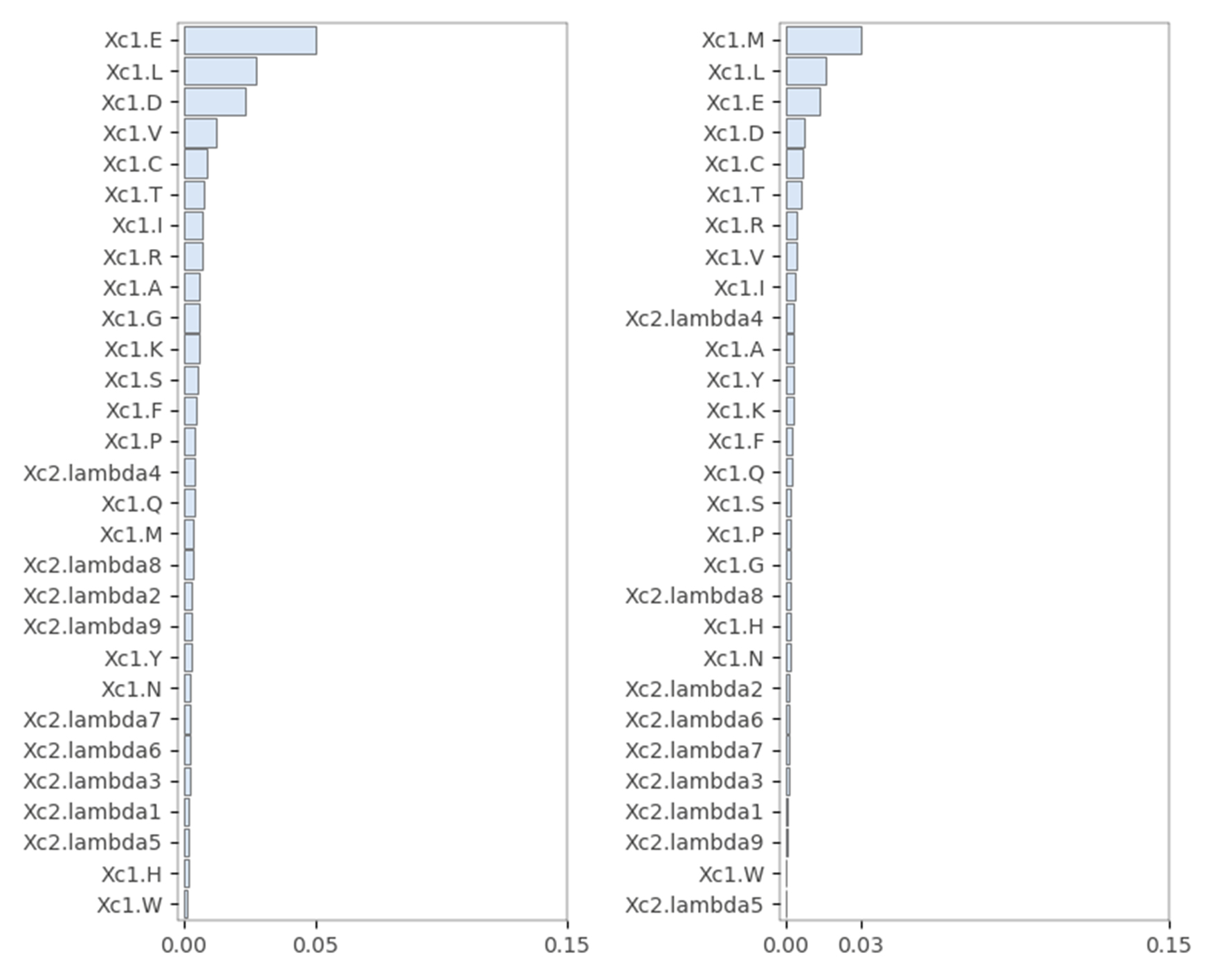
The PAAC feature: start codons “M” were deleted from protein sequences in the left image, and no amino acid was deleted in the right image
Training of “RF + PAAC + without M” (tree num 143):
accuracy: 0.944375
precision: 0.8883248730964467
recall: 0.8860759493670886
f1: 0.8871989860583016
Out-of-bag accuracy: 0.9394149933995692
Out-of-bag balanced accuracy: 0.92310748007037
AUC Score:0.9844550659173277
Training of “RF + PAAC + without M”: prediction accuracy of 2021 AMP sequences was 91.54%
Training of “RF + PAAC + without M”: prediction accuracy of 20000 non-AMP sequences (without M) was 97.16%
Training of “RF + PAAC + without M”: prediction accuracy of 20000 non-AMP sequences (with M) was 96.865%、
Training of “RF + CTDD + without M” (tree num 124):
accuracy: 0.91875
precision: 0.9187817258883249
recall: 0.9164556962025316
f1: 0.917617237008872
Out-of-bag accuracy:0.9309050465730572
Out-of-bag balanced accuracy:0.9309018454851675
AUC Score: 0.97790279731208
Training of “RF + CTDD + without M”: prediction accuracy of 2021 AMP sequences was 93.47%
Training of “RF + CTDD + without M”: prediction accuracy of 20000 AMP sequences (without M) was 93.25%
Training of “RF + CTDD + without M”: prediction accuracy of 20000 AMP sequences (with M) was 79.65%
Training of “RF + CTDD” (tree num 112):
accuracy: 0.96625
precision: 0.9659367396593674
recall: 0.9682926829268292
f1: 0.9671132764920829
Out-of-bag accuracy:0.9559293757820103
Out-of-bag balanced accuracy:0.9559898118461835
AUC Score: 0.9905315822388994
Training of “RF + CTDD”: prediction accuracy of 2021 AMP sequences was 96.98%
Training of “RF + CTDD”: prediction accuracy of 20000 AMP sequences (without M) was 67.02%
Training of “RF + CTDD”: prediction accuracy of 20000 AMP sequences (with M) was 94.92%
Training of “RF+PAAC” (tree num 126):
accuracy: 0.95125
precision: 0.9116883116883117
recall: 0.8886075949367088
f1: 0.8999999999999999
Out-of-bag accuracy:0.9446953380115334
Out-of-bag balanced accuracy:0.9307968041532002
AUC Score: 0.9862219654393614
Training of “RF+PAAC”: prediction accuracy of 2021 AMP sequences was 92.73%
Training of “RF+PAAC”: prediction accuracy of 20000 non-AMP sequences (without M) was 93.98%
Training of “RF+PAAC”: prediction accuracy of 20000 non-AMP sequences (with M) was 97.24%.