MATLAB Code for abnormal detection using SVDD
Version 2.2, 13-MAY-2022
Email: iqiukp@outlook.com
- SVDD model for one-class or binary classification
- Multiple kinds of kernel functions (linear, gaussian, polynomial, sigmoid, laplacian)
- Visualization of decision boundaries for 2D or 3D data
- Parameter optimization using Bayesian optimization, genetic algorithm, and pParticle swarm optimization
- Weighted SVDD model
- Hybrid-kernel SVDD model (K =w1×K1+w2×K2+...+wn×Kn)
- This version of this code is not compatible with the versions lower than R2016b.
- The label must be 1 for positive sample or -1 for negative sample.
- Detailed applications please see the provided demonstrations.
- This code is for reference only.
Please see the demonstration 📝 demo_BasicSVDD.m for details.
% generate dataset
ocdata = BinaryDataset();
ocdata.generate;
[trainData, trainLabel, testData, testLabel] = ocdata.partition;
% set parameter
kernel = BaseKernel('type', 'gaussian', 'gamma', 0.04);
cost = 0.3;
svddParameter = struct('cost', cost,...
'kernelFunc', kernel);
% creat an SVDD object
svdd = BaseSVDD(svddParameter);
% train SVDD model
svdd.train(trainData, trainLabel);
% test SVDD model
results = svdd.test(testData, testLabel);BinaryDatasetis designed to validate the svdd model only, you can use your data and please be careful to keep the naming of variables consistent, e.g.trainData,trainLabel,testData, andtestLabel.- Specifically, if the data does not have labels, please change the inputs for training or testing to
svdd.train(trainData)andresults = svdd.test(testData).
A class named SvddOptimization is defined to optimized the parameters. First define an optimization setting structure, then add it to the svdd parameter structure.The parameter optimization of the polynomial kernel function can only be achieved by using Bayesian optimization.
Please see the demonstration 📝 demo_ParameterOptimization.m for details.
% optimization setting
optimization.method = 'bayes'; %
optimization.maxIteration = 20;
optimization.display = 'on';
% SVDD parameter
svddParameter = struct('cost', cost,...
'kernelFunc', kernel,...
'optimization', optimization);The full properties of optimization are
method: optimization methods, only supported for 'bayes', 'pso', and 'ga'.variableName: variables that are to be optimized, including 'cost', 'degree', 'offset', and 'gamma'.variableType: variable type, specified as 'real' (real variable), 'integer' (integer variable).lowerBound: lower bound of variables.upperBound: upper bound of variables.maxIteration: max iterations.points: size of group or seed.display: visualization, 'on' or 'off'.
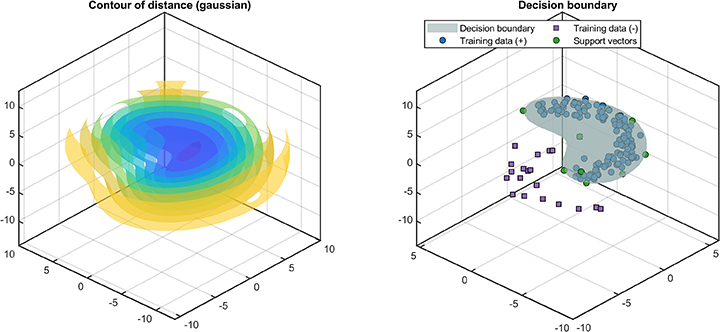
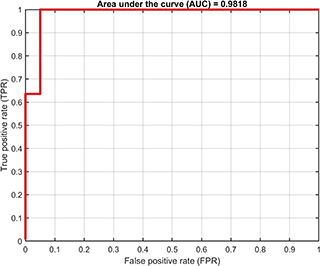
A class named SvddVisualization is defined to visualize the training and test results. Based on the trained SVDD model, the ROC curve of the training results (only supported for dataset containing both positive and negetive samples) is
% Visualization
svplot = SvddVisualization();
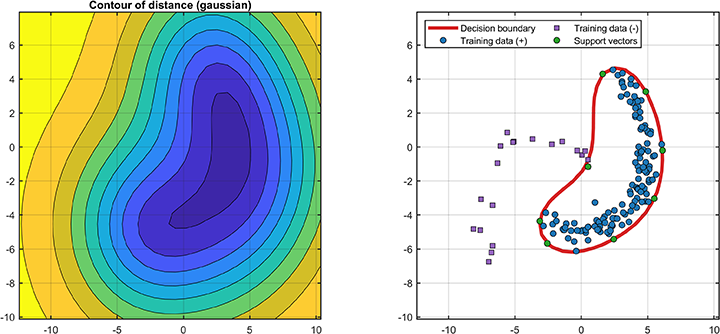
svplot.ROC(svdd);The decision boundaries (only supported for 2D/3D dataset) are
% Visualization
svplot = SvddVisualization();
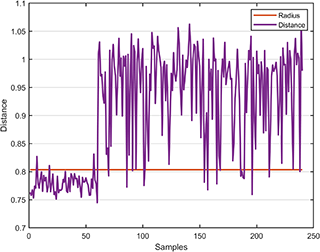
svplot.boundary(svdd);The distance between the test data and the hypersphere is
svplot.distance(svdd, results);A class named BinaryDataset is defined to generate and partition the 2D or 3D binary dataset.
Please see the demonstration 📝demo_BinaryDataset.m for details.
ocdata = BinaryDataset();
[data, label] = ocdata.generate;
[trainData, trainLabel, testData, testLabel] = ocdata.partition;The method generate is designed to generate dataset. The syntax of generate is
ocdata.generate;
data = ocdata.generate;
[data, label] = ocdata.generate;The method partition is designed to partition dataset into training dataset and test dataset. The syntax of partition is
[trainData, trainLabel, testData, testLabel] = ocdata.partition;The full Name-Value Arguments of class BinaryDataset are
shape: shape of dataset, 'banana' or 'circle'.dimensionality: dimensionality of dataset, 2 or 3.number: number of samples per class, for example: [200, 200].display: visualization, 'on' or 'off'.noise: noise added to dataset with range [0, 1]. For example: 0.2.ratio: ratio of the test set with range (0, 1). For example: 0.3.
A class named BaseKernel is defined to compute kernel function matrix.
Please see the demonstration 📝demo_KernelFunction.m for details.
%{
type -
linear : k(x,y) = x'*y
polynomial : k(x,y) = (γ*x'*y+c)^d
gaussian : k(x,y) = exp(-γ*||x-y||^2)
sigmoid : k(x,y) = tanh(γ*x'*y+c)
laplacian : k(x,y) = exp(-γ*||x-y||)
degree - d
offset - c
gamma - γ
%}
kernel = BaseKernel('type', 'gaussian', 'gamma', value);
kernel = BaseKernel('type', 'polynomial', 'degree', value);
kernel = BaseKernel('type', 'linear');
kernel = BaseKernel('type', 'sigmoid', 'gamma', value);
kernel = BaseKernel('type', 'laplacian', 'gamma', value);In this code, two cross-validation methods are supported: 'K-Folds' and 'Holdout'. For example, the cross-validation of 5-Folds is
svddParameter = struct('cost', cost,...
'kernelFunc', kernel,...
'KFold', 5);For example, the cross-validation of the Holdout method with a ratio of 0.3 is
svddParameter = struct('cost', cost,...
'kernelFunc', kernel,...
'Holdout', 0.3);For example, reducing the data to 2 dimensions can be set as
% SVDD parameter
svddParameter = struct('cost', cost,...
'kernelFunc', kernel,...
'PCA', 2);Please see the demonstration 📝demo_demo_DimReduPCA.m for details.
Notice: you only need to set PCA in svddParameter, and you don't need to process training data and test data separately.
An Observation-weighted SVDD is supported in this code.
Please see the demonstration 📝demo_ObservationWeight.m for details.
weight = rand(size(trainData, 1), 1);
% SVDD parameter
svddParameter = struct('cost', cost,...
'kernelFunc', kernel,...
'weight', weight);Notice: the size of 'weigh' should be m×1, where m is the number of training samples.
A demo for SVDD using Hybrid kernel functions (K =w1×K1+w2×K2+...+wn×Kn).
Please see the demonstration 📝demo_HybridKernelSVDD.m for details.
kernel_1 = BaseKernel('type', 'gaussian', 'gamma', 1);
kernel_2 = BaseKernel('type', 'polynomial', 'degree', 3);
kernelWeight = [0.5, 0.5];
cost = 0.9;
svddParameter = struct('cost', cost,...
'kernelFunc', [kernel_1, kernel_2],...
'kernelWeight', kernelWeight);