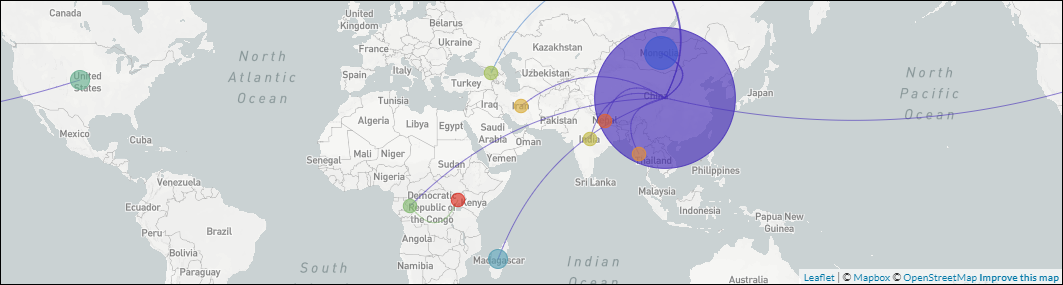
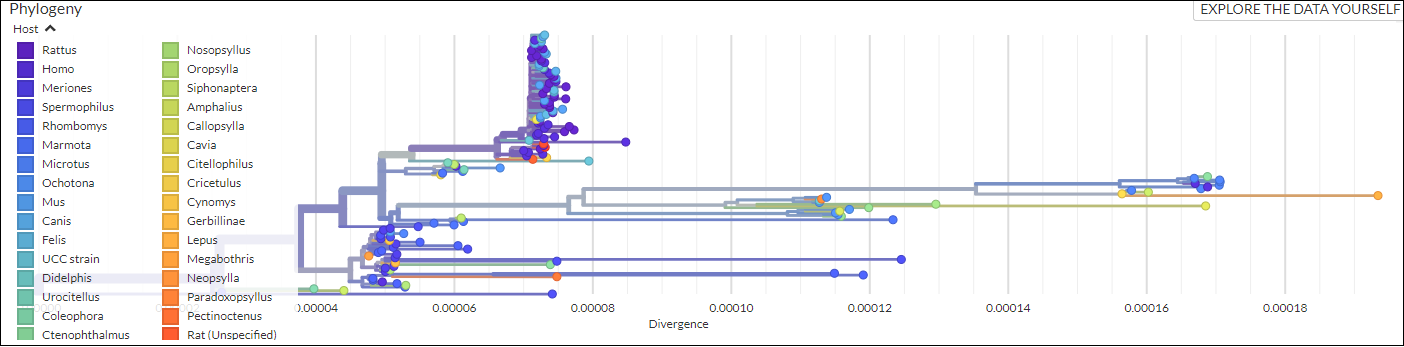
An open-source pipeline to construct a global phylogeny of the plague pathogen Yersinia pestis.
- Create a metadata database of NCBI genomic assemblies and SRA data.
- Download assemblies and SRA fastq files.
- Align assemblies and fastq files to a reference genome.
- Mask problematic regions.
- Evaluate statistics.
- Estimate a maximum-likelihood phylogeny.
- Estimate a time-scaled phylogeny.
- Web-based visualization.
- Presenting “The Plague”: Digital Exhibits as Interdisciplinary Method.
DHSI Conference and Colloquium. June 5, 2020.
Katherine Eaton, Nukhet Varlik, Ann Carmichael, Brian Golding, Hendrik Poinar.
Digital Exhibit • Talk
- Plagues of the Past and Present.
Lewis & Ruth Sherman Centre for Digital Scholarship. June 2, 2020.
Katherine Eaton
Digital Exhibit • Blog Post 1 • Blog Post 2 *
All install options start by cloning the pipeline repo.
git clone https://github.com/ktmeaton/plague-phylogeography.git
cd plague-phylogeography- Create a conda environment with dependencies.
- Mamba is not strictly necessary but it is heavily recommended.
conda install -c conda-forge mamba
mamba env create -f workflow/envs/merge/environment.yaml- Install LSD2 (not available through conda).
# Download binary
wget https://github.com/tothuhien/lsd2/releases/download/v1.9.9/lsd2_unix
# Move into the conda binary path and rename
mv lsd2_unix ~/miniconda3/envs/plague-phylogeography/bin/lsd2- Test the help command.
snakemake --profile profiles/laptop help(While mamba is not strictly necessary, it is heavily recommended.)
snakemake --profile profiles/laptop alldocker pull ktmeaton/plague-phylogeography:dev
docker run \
-v $PWD:/pipeline \
-w /pipeline \
ktmeaton/plague-phylogeography:dev \
snakemake --profile profiles/laptop helpsnakemake --profile profiles/laptop alldocker run \
-v $PWD:/pipeline \
-w /pipeline \
ktmeaton/plague-phylogeography:dev \
snakemake --profile profiles/laptop allAuthor: Katherine Eaton
Logo: Emil Karpinski, Katherine Eaton