The HistFactory p.d.f. template [CERN-OPEN-2012-016] is per-se independent of its implementation in ROOT and sometimes, it’s useful to be able to run statistical analysis outside of ROOT, RooFit, RooStats framework.
This repo is a pure-python implementation of that statistical model for multi-bin histogram-based analysis and its interval estimation is based on the asymptotic formulas of “Asymptotic formulae for likelihood-based tests of new physics” [arXiv:1007.1727]. The aim is also to support modern computational graph libraries such as PyTorch and TensorFlow in order to make use of features such as autodifferentiation and GPU acceleration.
For an in depth walkthrough of usage of the latest release of pyhf visit the pyhf tutorial.
This is how you use the pyhf Python API to build a statistical model and run basic inference:
>>> import pyhf
>>> pyhf.set_backend("numpy")
>>> model = pyhf.simplemodels.uncorrelated_background(
... signal=[12.0, 11.0], bkg=[50.0, 52.0], bkg_uncertainty=[3.0, 7.0]
... )
>>> data = [51, 48] + model.config.auxdata
>>> test_mu = 1.0
>>> CLs_obs, CLs_exp = pyhf.infer.hypotest(
... test_mu, data, model, test_stat="qtilde", return_expected=True
... )
>>> print(f"Observed: {CLs_obs:.8f}, Expected: {CLs_exp:.8f}")
Observed: 0.05251497, Expected: 0.06445321Alternatively the statistical model and observational data can be read from its serialized JSON representation (see next section).
>>> import pyhf
>>> import requests
>>> pyhf.set_backend("numpy")
>>> url = "https://raw.githubusercontent.com/scikit-hep/pyhf/main/docs/examples/json/2-bin_1-channel.json"
>>> wspace = pyhf.Workspace(requests.get(url).json())
>>> model = wspace.model()
>>> data = wspace.data(model)
>>> test_mu = 1.0
>>> CLs_obs, CLs_exp = pyhf.infer.hypotest(
... test_mu, data, model, test_stat="qtilde", return_expected=True
... )
>>> print(f"Observed: {CLs_obs:.8f}, Expected: {CLs_exp:.8f}")
Observed: 0.35998409, Expected: 0.35998409Finally, you can also use the command line interface that pyhf provides
$ cat << EOF | tee likelihood.json | pyhf cls
{
"channels": [
{ "name": "singlechannel",
"samples": [
{ "name": "signal",
"data": [12.0, 11.0],
"modifiers": [ { "name": "mu", "type": "normfactor", "data": null} ]
},
{ "name": "background",
"data": [50.0, 52.0],
"modifiers": [ {"name": "uncorr_bkguncrt", "type": "shapesys", "data": [3.0, 7.0]} ]
}
]
}
],
"observations": [
{ "name": "singlechannel", "data": [51.0, 48.0] }
],
"measurements": [
{ "name": "Measurement", "config": {"poi": "mu", "parameters": []} }
],
"version": "1.0.0"
}
EOFwhich should produce the following JSON output:
{
"CLs_exp": [
0.0026062609501074576,
0.01382005356161206,
0.06445320535890459,
0.23525643861460702,
0.573036205919389
],
"CLs_obs": 0.05251497423736956
}- Implemented variations:
- ☑ HistoSys
- ☑ OverallSys
- ☑ ShapeSys
- ☑ NormFactor
- ☑ Multiple Channels
- ☑ Import from XML + ROOT via uproot
- ☑ ShapeFactor
- ☑ StatError
- ☑ Lumi Uncertainty
- ☑ Non-asymptotic calculators
- Computational Backends:
- ☑ NumPy
- ☑ PyTorch
- ☑ TensorFlow
- ☑ JAX
- Optimizers:
- ☑ SciPy (
scipy.optimize) - ☑ MINUIT (
iminuit)
- ☑ SciPy (
All backends can be used in combination with all optimizers. Custom user backends and optimizers can be used as well.
- ☐ StatConfig
results obtained from this package are validated against output computed from HistFactory workspaces
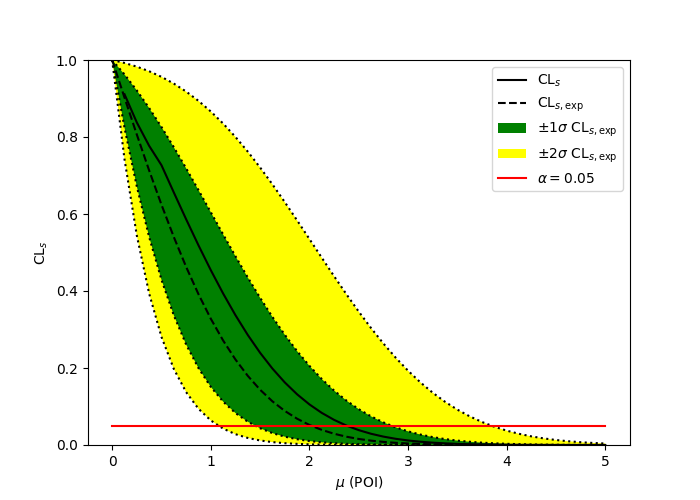
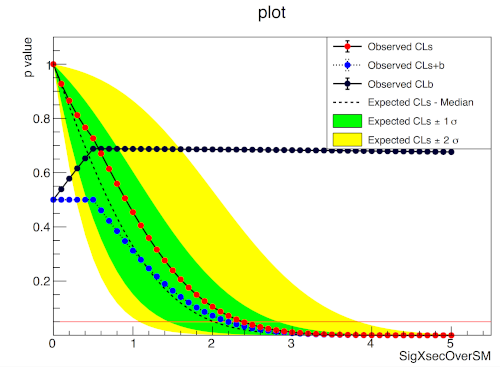
import pyhf
import numpy as np
import matplotlib.pyplot as plt
from pyhf.contrib.viz import brazil
pyhf.set_backend("numpy")
model = pyhf.simplemodels.uncorrelated_background(
signal=[10.0], bkg=[50.0], bkg_uncertainty=[7.0]
)
data = [55.0] + model.config.auxdata
poi_vals = np.linspace(0, 5, 41)
results = [
pyhf.infer.hypotest(
test_poi, data, model, test_stat="qtilde", return_expected_set=True
)
for test_poi in poi_vals
]
fig, ax = plt.subplots()
fig.set_size_inches(7, 5)
brazil.plot_results(poi_vals, results, ax=ax)
fig.show()pyhf
ROOT
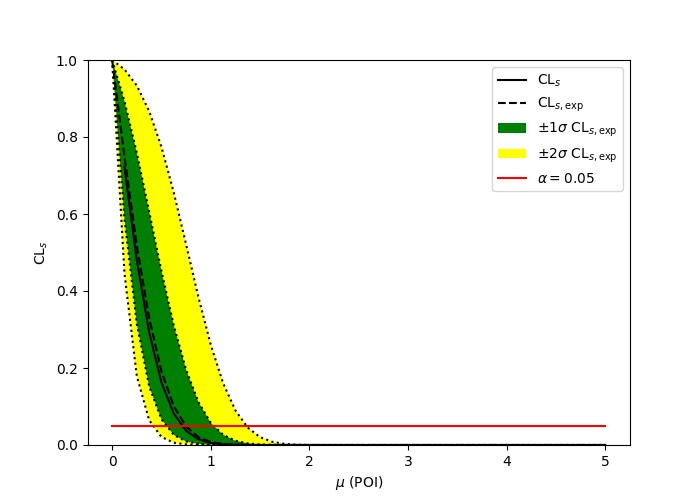
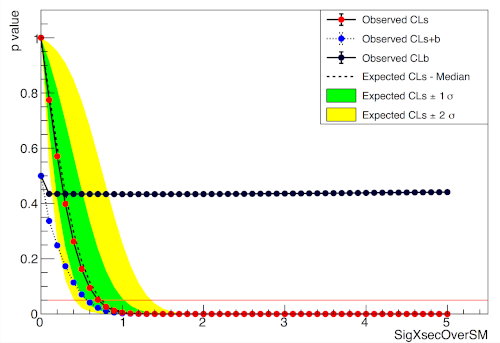
import pyhf
import numpy as np
import matplotlib.pyplot as plt
from pyhf.contrib.viz import brazil
pyhf.set_backend("numpy")
model = pyhf.simplemodels.uncorrelated_background(
signal=[30.0, 45.0], bkg=[100.0, 150.0], bkg_uncertainty=[15.0, 20.0]
)
data = [100.0, 145.0] + model.config.auxdata
poi_vals = np.linspace(0, 5, 41)
results = [
pyhf.infer.hypotest(
test_poi, data, model, test_stat="qtilde", return_expected_set=True
)
for test_poi in poi_vals
]
fig, ax = plt.subplots()
fig.set_size_inches(7, 5)
brazil.plot_results(poi_vals, results, ax=ax)
fig.show()pyhf
ROOT
To install pyhf from PyPI with the NumPy backend run
python -m pip install pyhfand to install pyhf with all additional backends run
python -m pip install pyhf[backends]or a subset of the options.
To uninstall run
python -m pip uninstall pyhfFor model specification, API reference, examples, and answers to FAQs visit the pyhf documentation.
If you have a question about the use of pyhf not covered in the
documentation, please ask a question
on the GitHub Discussions.
If you believe you have found a bug in pyhf, please report it in the
GitHub
Issues.
If you're interested in getting updates from the pyhf dev team and release
announcements you can join the pyhf-announcements mailing list.
As noted in Use and Citations,
the preferred BibTeX entry for citation of pyhf includes both the
Zenodo archive and the
JOSS paper:
@software{pyhf,
author = {Lukas Heinrich and Matthew Feickert and Giordon Stark},
title = "{pyhf: v0.7.5}",
version = {0.7.5},
doi = {10.5281/zenodo.1169739},
url = {https://doi.org/10.5281/zenodo.1169739},
note = {https://github.com/scikit-hep/pyhf/releases/tag/v0.7.5}
}
@article{pyhf_joss,
doi = {10.21105/joss.02823},
url = {https://doi.org/10.21105/joss.02823},
year = {2021},
publisher = {The Open Journal},
volume = {6},
number = {58},
pages = {2823},
author = {Lukas Heinrich and Matthew Feickert and Giordon Stark and Kyle Cranmer},
title = {pyhf: pure-Python implementation of HistFactory statistical models},
journal = {Journal of Open Source Software}
}pyhf is openly developed by Lukas Heinrich, Matthew Feickert, and Giordon Stark.
Please check the contribution statistics for a list of contributors.
- 2022-09-12: 2000 GitHub issues and pull requests. (See PR #2000)
- 2021-12-09: 1000 commits to the project. (See PR #1710)
- 2020-07-28: 1000 GitHub issues and pull requests. (See PR #1000)
Matthew Feickert has received support to work on pyhf provided by NSF
cooperative agreements OAC-1836650
and PHY-2323298 (IRIS-HEP)
and grant OAC-1450377 (DIANA/HEP).
pyhf is a NumFOCUS Affiliated Project.