Meet-EU Team SB2 - Link to MEET-EU website
Topic B : Chromosome compartments
Project of the MEET-U course for our university Sorbonne Université
Meetings : One meeting each week every wednesday afternoon from 4pm to 6pm
Responsability list :
| Role | Name |
|---|---|
| Manager Expert | Rouquaya Mouss |
| Technician Expert | Damien Legros & Cédric Cornede |
| Scientific Expert | Hamid Hachemi |
| Delivrable Expert | Arnaud Quelin |
First you need to clone our repository :
git clone https://github.com/meet-eu-21/Team-SB2
After that you have to install the requirements :
pip install -r requirements.txt
Then to launch the code :
python main.py
The following panel will come up, you can enter the numbers of the chromosomes you want to analyse and select the cell lines and the resolution (be careful ! : launching too many chromosomes can make a runtime memory error).
Here you have an example of the selection to run an analysis for GM12878, HUVEC and IMR90 with the chromosomes 11, X and 4 in 100kb.
The notebook Plot_histo.pynb can then be used to make the histograms of the results to compare the cell lines.
Class diagram of our code :
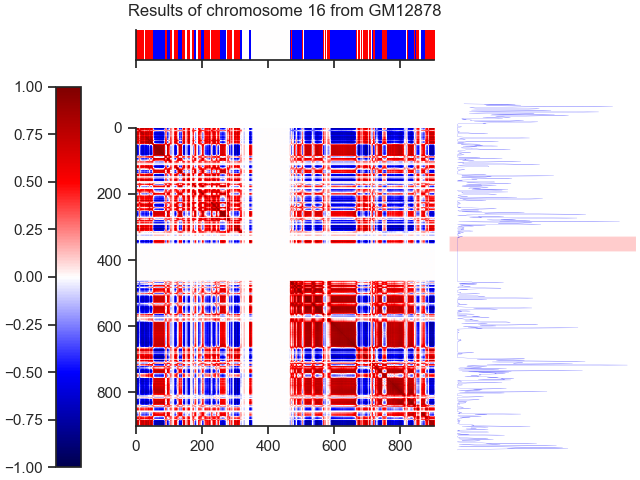
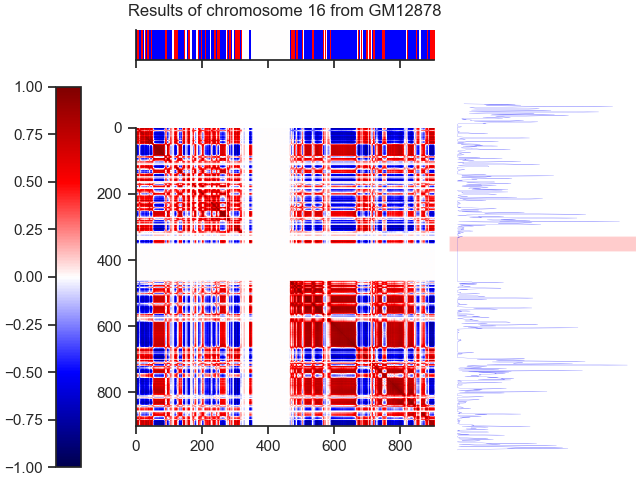
Our results for 100kb are available in the folder /Results
All .py files except color_b.py are used for our analysis. Two methods are used :
- With the first method, we search the number of compartments with an HMM using the data from the correlation matrix
- With the second method, we search the number of compartments with an HMM using the data from epigenetic marks.
The file color_b.py can be used with Pymol to see the colors of the compartments of the .pdb files of our results. (color_b.py can be put in Pymol folder, no need of the path before color_b.py in the following command if that's the case). You need to run the following commands in Pymol after opening the .pdb result you want to see :
run path_to_our_folder\Team-SB2\color_b.py
color_b()
Here is an example of the colors that can be seen on the .pdb file of chromosome 22 of HMEC with the compartments of the HMM method with correlation matrix :

###################################HMM Contact###################################
Similarity score with Leopold: 74.972 %
Best compartment number : 7
###################################HMM Epigenetic###################################
Similarity score with Leopold: 64.673 %
Best compartment number : 5