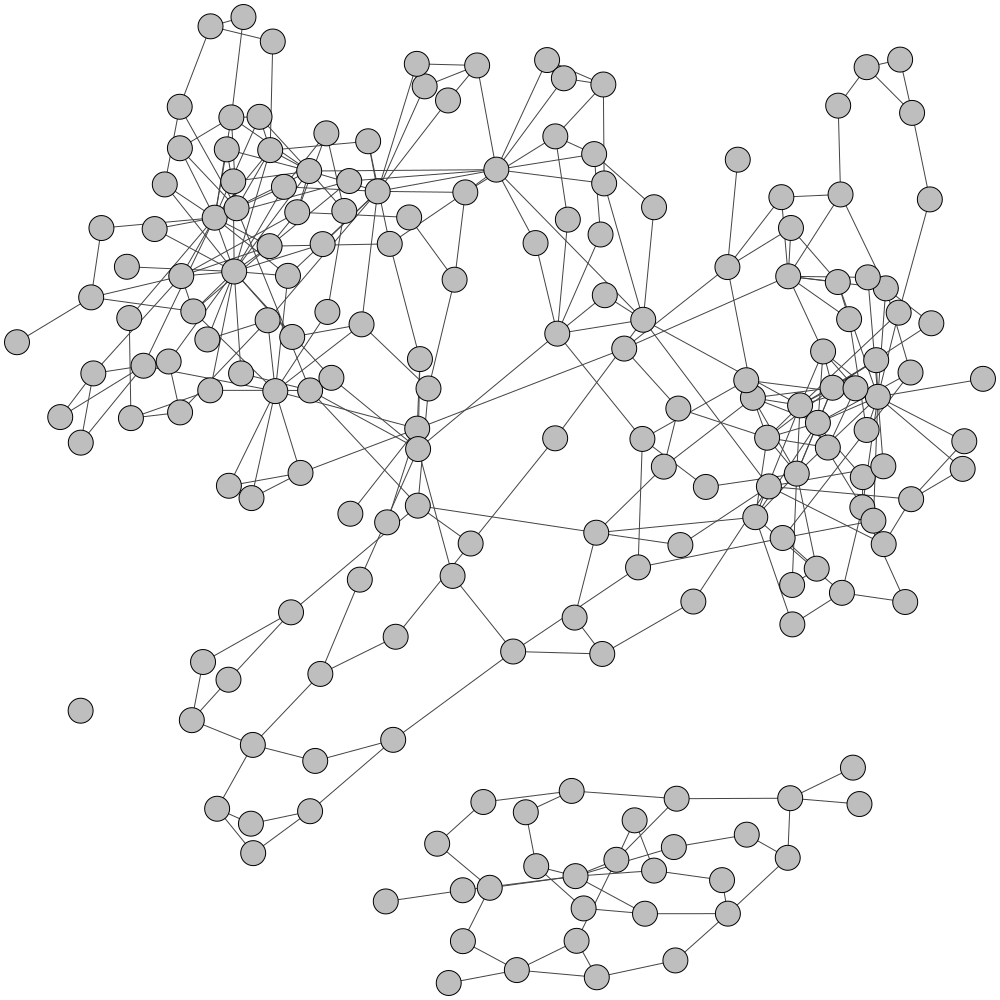
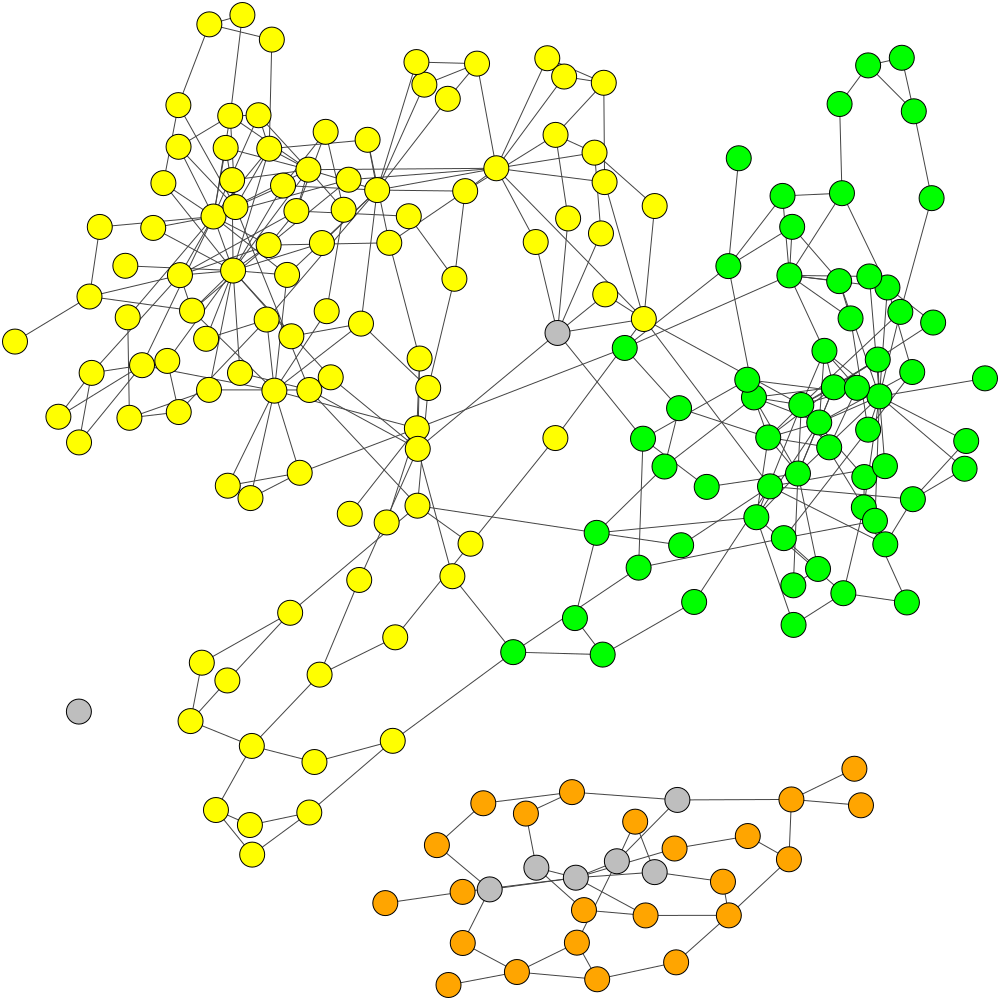
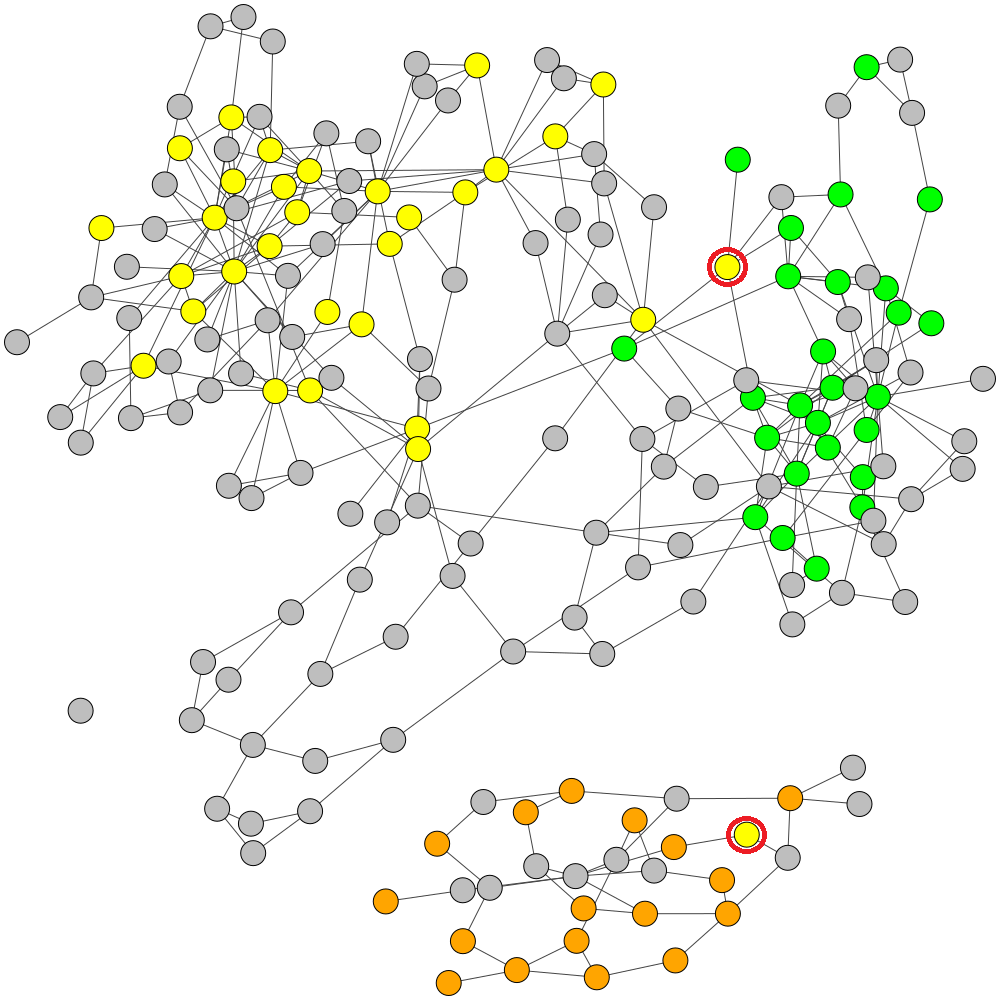
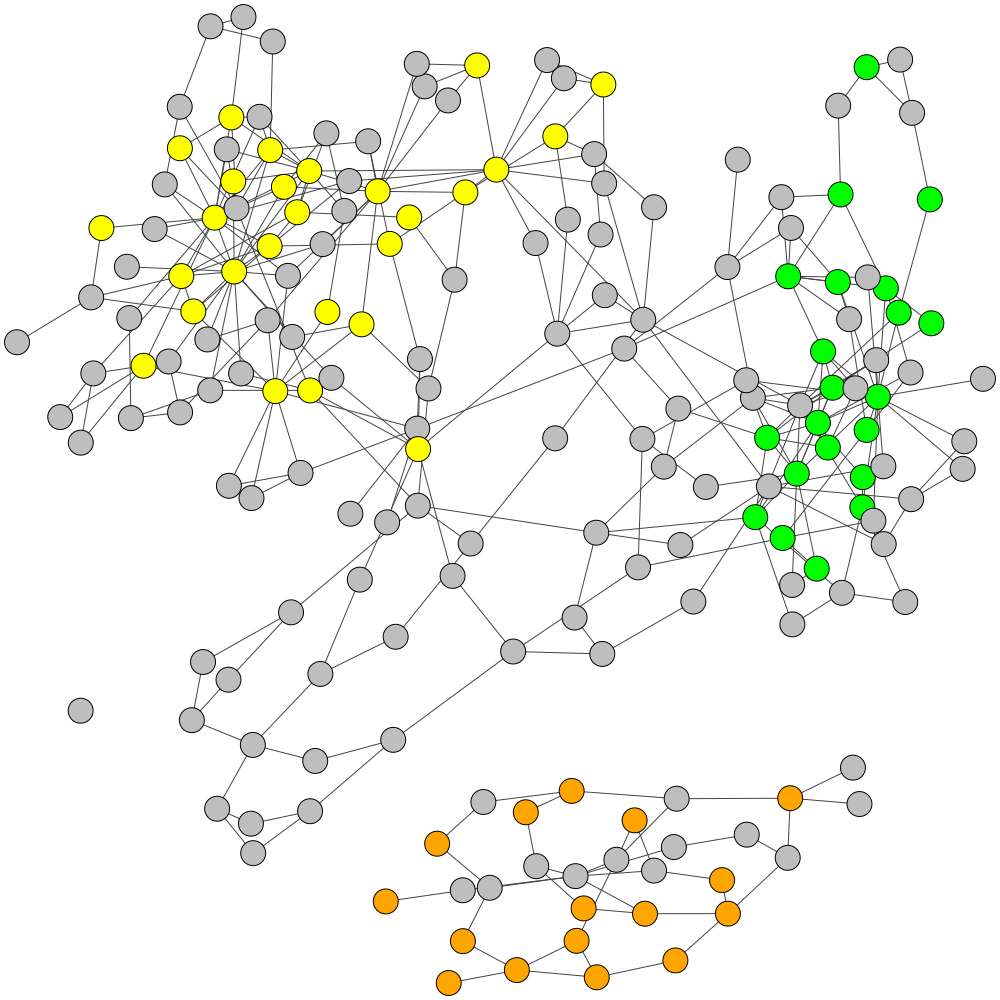
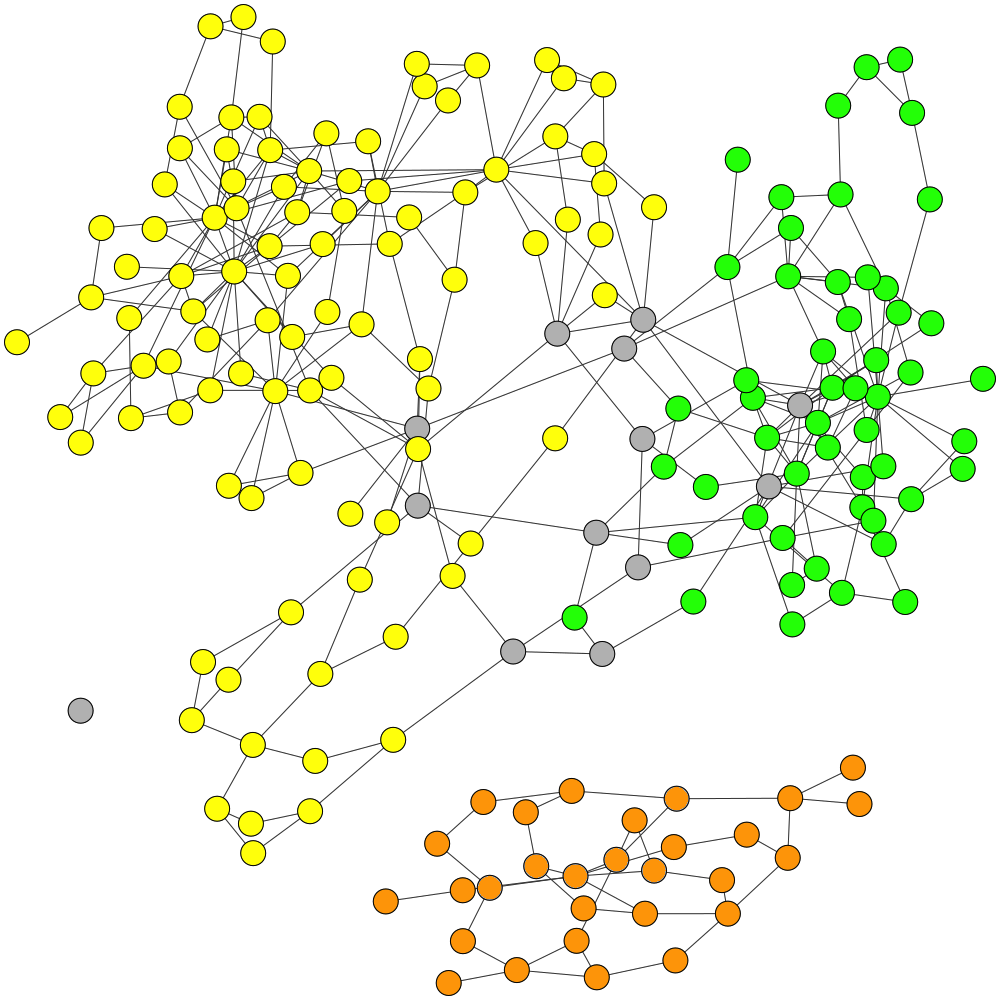
GraphBin is an NGS data-based metagenomic contig bin refinement tool that makes use of the contig connectivity information from the assembly graph to bin contigs. It utilizes the binning result of an existing binning tool and a label propagation algorithm to correct mis-binned contigs and predict the labels of contigs which are discarded due to short length.
For detailed instructions on installation, usage and visualisation, please refer to the documentation hosted at Read the Docs.
GraphBin installation requires python 3 to run. The following dependencies are required to run GraphBin and related support scripts.
You can install GraphBin using the bioconda distribution. You can download
Anaconda or Miniconda which contains conda.
# add channels
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forge
# create conda environment
conda create -n graphbin
# activate conda environment
conda activate graphbin
# install graphbin
conda install -c bioconda graphbin
# check graphbin installation
graphbin -h
You can install GraphBin using pip from the PyPI distribution.
pip install graphbin
For development purposes, please clone the repository and install via flit.
# clone repository to your local machine
git clone https://github.com/metagentools/GraphBin.git
# go to repo directory
cd GraphBin
# install flit
pip install flit
# install graphbin via flit
flit install -s --python `which python`
# SPAdes version
graphbin --assembler spades --graph /path/to/graph_file.gfa --contigs /path/to/contigs.fasta --paths /path/to/paths_file.paths --binned /path/to/binning_result.csv --output /path/to/output_folder
# SGA version
graphbin --assembler sga --graph /path/to/graph_file.asqg --contigs /path/to/contigs.fa --binned /path/to/binning_result.csv --output /path/to/output_folder
# MEGAHIT version
graphbin --assembler megahit --graph /path/to/graph_file.gfa --contigs /path/to/contigs.fa --binned /path/to/binning_result.csv --output /path/to/output_folder
If you use GraphBin in your work, please cite GraphBin as,
Vijini Mallawaarachchi, Anuradha Wickramarachchi, Yu Lin. GraphBin: Refined binning of metagenomic contigs using assembly graphs. Bioinformatics, Volume 36, Issue 11, June 2020, Pages 3307–3313, DOI: https://doi.org/10.1093/bioinformatics/btaa180
@article{10.1093/bioinformatics/btaa180,
author = {Mallawaarachchi, Vijini and Wickramarachchi, Anuradha and Lin, Yu},
title = "{GraphBin: refined binning of metagenomic contigs using assembly graphs}",
journal = {Bioinformatics},
volume = {36},
number = {11},
pages = {3307-3313},
year = {2020},
month = {03},
abstract = "{The field of metagenomics has provided valuable insights into the structure, diversity and ecology within microbial communities. One key step in metagenomics analysis is to assemble reads into longer contigs which are then binned into groups of contigs that belong to different species present in the metagenomic sample. Binning of contigs plays an important role in metagenomics and most available binning algorithms bin contigs using genomic features such as oligonucleotide/k-mer composition and contig coverage. As metagenomic contigs are derived from the assembly process, they are output from the underlying assembly graph which contains valuable connectivity information between contigs that can be used for binning. We propose GraphBin, a new binning method that makes use of the assembly graph and applies a label propagation algorithm to refine the binning result of existing tools. We show that GraphBin can make use of the assembly graphs constructed from both the de Bruijn graph and the overlap-layout-consensus approach. Moreover, we demonstrate improved experimental results from GraphBin in terms of identifying mis-binned contigs and binning of contigs discarded by existing binning tools. To the best of our knowledge, this is the first time that the information from the assembly graph has been used in a tool for the binning of metagenomic contigs. The source code of GraphBin is available at https://github.com/Vini2/GraphBin.vijini.mallawaarachchi@anu.edu.au or yu.lin@anu.edu.auSupplementary data are available at Bioinformatics online.}",
issn = {1367-4803},
doi = {10.1093/bioinformatics/btaa180},
url = {https://doi.org/10.1093/bioinformatics/btaa180},
eprint = {https://academic.oup.com/bioinformatics/article-pdf/36/11/3307/33329097/btaa180.pdf},
}GraphBin is funded by an Essential Open Source Software for Science Grant from the Chan Zuckerberg Initiative.