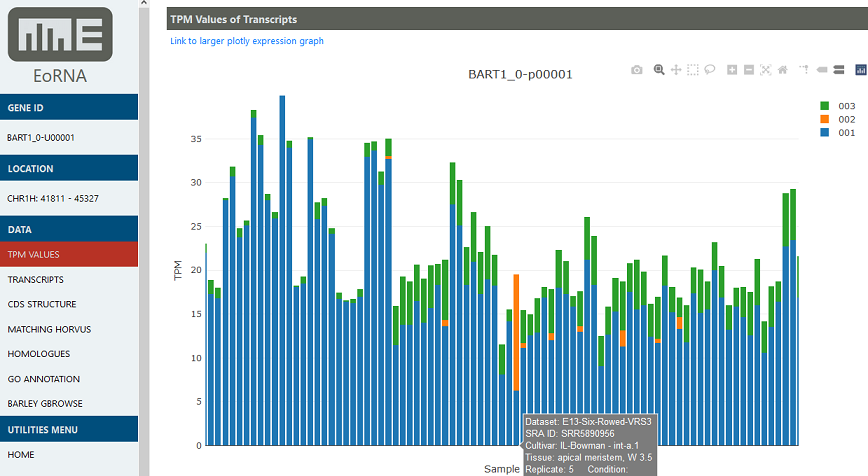
The EORNA website is designed to store and visualize gene and transcript abundance data on demand as transcripts per million (TPM) across all samples and all the genes within the barley genome. Abundances of alternative transcript expression for a gene are displayed as a Manhattan plot using Plotly.
EORNA provides access to the gene and transcript abundances quantified using data from 22 RNA-seq experiments, covering 843 separate samples, as well as the sequence data and annotation for the Barley Reference Transcript Dataset.
The EORNA website can be found at https://ics.hutton.ac.uk/eorna/index.html.
The paper accompanying the website and the data analysis is at Bioarxiv : EoRNA, a barley gene and transcript abundance database. Linda Milne, Micha Bayer, Paulo Rapazote-Flores, Claus-Dieter Mayer, Robbie Waugh, Craig G. Simpson doi: https://doi.org/10.1101/2020.11.24.395749