Provides necessary python code used in our paper: Breaking Medical Data Sharing Boundaries by Employing Artificial Radiographs.
- MS-SSIM histogram results is uploaded to
SSIM.Resultsfolder. - Add code for federated learning GAN at
Federated_GANfolder.
Artificial intelligence (AI) has the potential to change medicine fundamentally. Here, expert
knowledge provided by AI can enhance diagnosis by comprehensive image features. Unfortunately,
existing algorithms often stay behind expectations, as databases used for training are usually
too small, incomplete, and heterogeneous in quality. Additionally, data protection constitutes
a serious obstacle to data sharing. We propose to use generative models (GM) to produce
high-resolution artificial radiographs, which are free of personal identifying information. Blinded
analyses by computer vision and radiology experts proved the high similarity of artificial and real
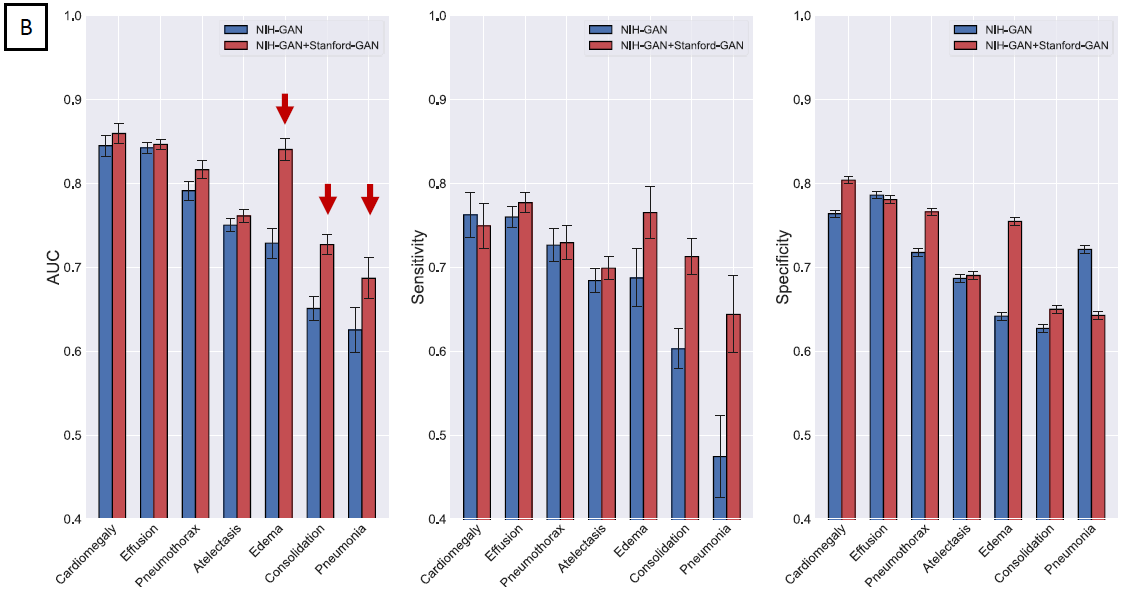
radiographs. The combination of pooled GM improves the performance of computer vision algorithms
trained on smaller datasets and the integration of artificial data into patient data repositories
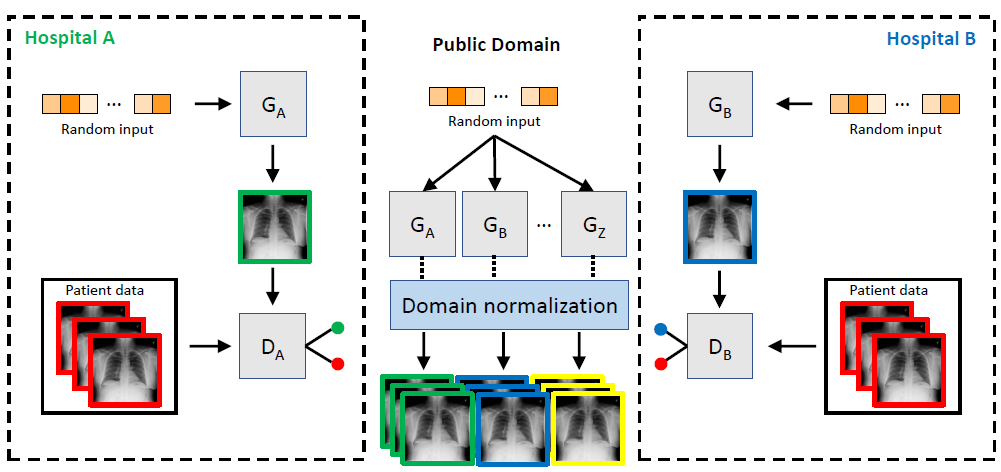
can compensate for underrepresented disease entities. By integrating federated learning strategies,
even small hospitals can participate in the training of GMs. We envision that our approach could
lead to scalable databases of anonymous medical images enabling standardized radiomic analyses
at multiple sites.
- TensorFlow 1.9.0
- Pytorch 1.1.0
- Image preprocessing
python -m preprocess.CheXpert_process
- CheXpert label generation
preprocess.Stanford_CSV.ipynb
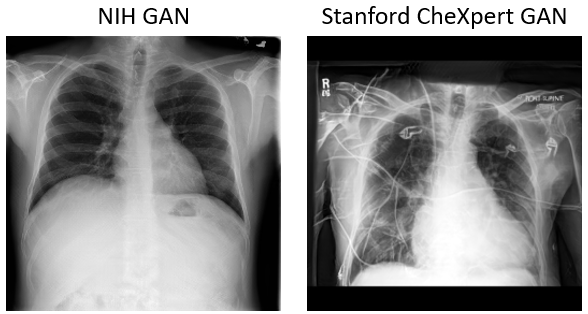
Artificial radiographs used in this paper is generated by progressive growing GAN. The GAN part of this repository is modified from the official implementation of Nvidia. Artificial radiographs can prepared by run the following:
- prepare
*.tfrecordsfiles:
python dataset_tool.py create_from_images_labels [output Dir] [input image Dir] [label Dir] [label file]
such as:
python dataset_tool.py create_from_images_labels /media/tianyu.han/mri-scratch/DeepLearning/dataset/CheXpert_256 /media/tianyu.han/mri-scratch/DeepLearning/CheXpert_Dataset/images/ /media/tianyu.han/mri-scratch/DeepLearning/CheXpert_Dataset/labels/ label_CheXpert.npy
- Edit
config.pyto specify the dataset and training configuration by uncommenting/editing specific lines. - Run the training script with
python train.py - After training is converged, one can use
ProGAN.reproduce_trainset.ipynbto produce artificial radiographs - The trained NIH and CheXpert GAN models can be downloaded from this link.
- For evaluating the training of GANs, one can compute Frechet Inception Distance and MS-SSIM by using
ProGAN.metrics.frechet_inception_distance.pyandSSIM.ms_ssim.py
Inspired by the previous work of CheXnet, we trained a DenseNet-121 (ImageNet weights) classifier to evaluate the performance of generated radiographs. The code of classifier is contained in folder Thorax Classifier.s
- modify the path in
retrain.pyand simply runpython retrain.py - All used classifier models can be downloaded via this link.
- To obtain the confidence interval, one can use bootstrapping method by run
python -m Analysis.bootstrapping.
For each pathology, 5,000 random artificial radiographs with a pathology label drawn from a uniform distribution
between 0.0 and 1.0 were generated. The images were then rated by the classifier network and Pearson’s correlation
coefficient was calculated for each pairing of pathologies.
The experimental code is uploaded to folder Correlation to reproduce our findings.
- Our correlation results can be found at here.
- To make this approach applicable, domain adaptation techniques such as Cycle-GAN based image translation is also consider in this study. We successfully trained a Cycle-GAN to achieve NIH and CheXpert style transformation.
- The pretrained NIH2CheXpert Cycle-GAN model can be downloaded here.
If you use this code for your research, please cite our papers.
@article{han2019breaking,
title={Breaking Medical Data Sharing Boundaries by Employing Artificial Radiographs},
author={Han, Tianyu and Nebelung, Sven and Haarburger, Christoph and Horst, Nicolas and Reinartz, Sebastian and Merhof, Dorit and Kiessling, Fabian and Schulz, Volkmar and Truhn, Daniel},
journal={BioRxiv},
pages={841619},
year={2019},
publisher={Cold Spring Harbor Laboratory}
}
This project is licensed under the MIT License - see the LICENSE.md file for details
- Official progressive growing GAN code in TensorFlow: https://github.com/tkarras/progressive_growing_of_gans/blob/master/README.md
- Official Cycle-GAN code in Pytorch: https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix
- The Stanford Machine Learning Group: https://stanfordmlgroup.github.io/