Warning
SPREAD hosting on AWS will be discontinued at the end of year 2025 due to funding issues. For other phylogeographic visualisation options, please visit https://beast.community/spread4.
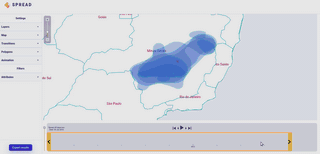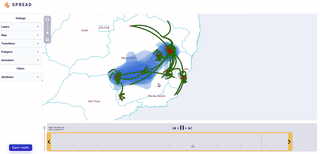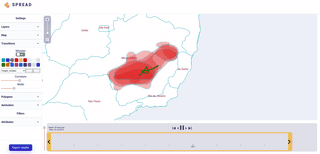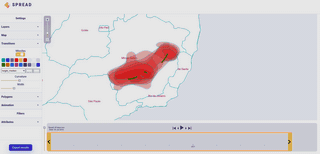
Spread vizualizes how viruses and other pathogens are spreading in time and space. It creates shareable, interactive and time-animated vizualisation.
@article{10.1093/ve/veac088,
author = {Nahata, Kanika D and Bielejec, Filip and Monetta, Juan and Dellicour, Simon and Rambaut, Andrew and Suchard, Marc A and Baele, Guy and Lemey, Philippe},
title = "{SPREAD 4: online visualisation of pathogen phylogeographic reconstructions}",
journal = {Virus Evolution},
volume = {8},
number = {2},
year = {2022},
month = {09},
issn = {2057-1577},
doi = {10.1093/ve/veac088},
url = {https://doi.org/10.1093/ve/veac088},
note = {veac088},
eprint = {https://academic.oup.com/ve/article-pdf/8/2/veac088/46671630/veac088.pdf},
}To be able to easily pick up stack used to build spread, one should be familiar with following topics:
The diagram below presents an overview of the architecture of spread:
- API is a gateway service, exposing graphql endpoints and publishing messages to the SQS queue.
- Worker is a messaging service wrapping the phylogeographic tree graphs parsing library libspread, multiple workers compete for the messages published by the API.
- Relational Database and S3 object storage are used for persistance.
- User-facing interface facilitates communicating with the API from the client.
- Visualization engine accepts S3 stored output, animates and displays it on the geographical map.
Make sure you have tmux multiplexer installed.
Execute ./start_all_components in your terminal window and it will start all the components in separate tmux windows.
Make sure you have docker and docker-compose installed. You will also need maven and clojure cli-tool.
Source the environment variables and start the dev infrastructure:
source env/dev
docker-compose -f deployments/dev/docker-compose.yml upDeploy the database schema changes:
cd services/db-migration
source ../../env/dev
mvn package
mvn liquibase:updateCompile and package libspread:
mvn clean packageStart an instance of the worker-service:
clj -A:run-workerStart an instance of the api-service from the comand-line:
clj -A:run-apiStart an instance of the api-service from the REPL:
M+x cider-jack-in
C-c M-n-n api.main
(restart)
In the default dev environment a GraphQL IDE is started at:
http://127.0.0.1:3001/ide
Make sure you have yarn installed.
Install dependencies:
yarn depsStart watcher and local server
yarn watchOpen in browser: http://localhost:8020
To get the cljs REPL:
M+x cider-connect-cljsSelect localhost and the nREPL port printed by the watcher (e.g. 46000), select shadow and :ui as the build.
Start watcher and local server
yarn watch:viewerStart a watcher on the libspread test suite:
mvn fizzed-watcher:runStart a watcher on the spread's integration test suite:
bin/kaocha --watch
This project exists thanks to all the people who contribute.
MIT © Filip Bielejec