If you have ever felt at all disappointed seeing Ruby's computational performance particularly for large-scale scientific computation, VirtualModule may save you a little. It offers you a way to run your arbitrary Ruby code on the other language VM process (currently only Julia is supported). What make VirtualModule possible to do this is ruby2julia transpiler (which is at very early stage of development) and IPC using msgpack.
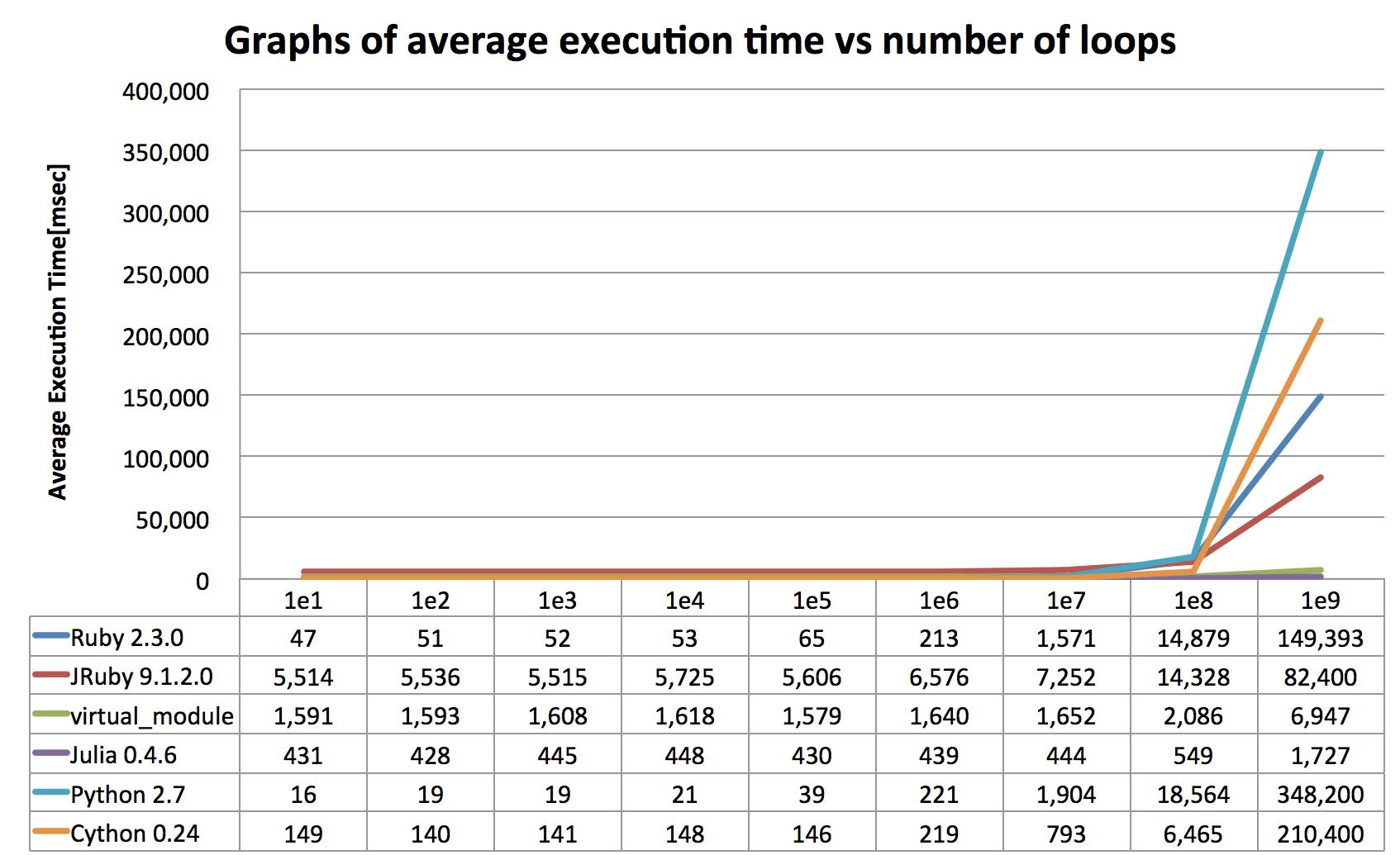
Let's take a look at results of benchmark program of which scores were taken on my MacBook Pro (OSX10.11, Core i5 2.9GHz, 8GB). Essentially this benchmarking program just repeats adding an integer value to floating point value cumulatively, which is represented as x=2.5; 1.upto(N){|i| x=x+i} in Ruby. If you run this program with setting of smaller number of loops (upto 10,000,000 times), VirtualModule doesn't make sense at all as you can see:
However as the number of loops gets bigger(upto 1,000,000,000 times), VirtualModule significantly reduces total execution time.
(For the sake of honor of python and cython, here is another benchmarking results measured on Ubuntu 16 on DigitalOcean (8CPUs, 16GB Mem). It records better score for both python2.7 and Cython than the one with my MacBook Pro, although I'm not sure why this happens. Does anyone can guess why this happens?)
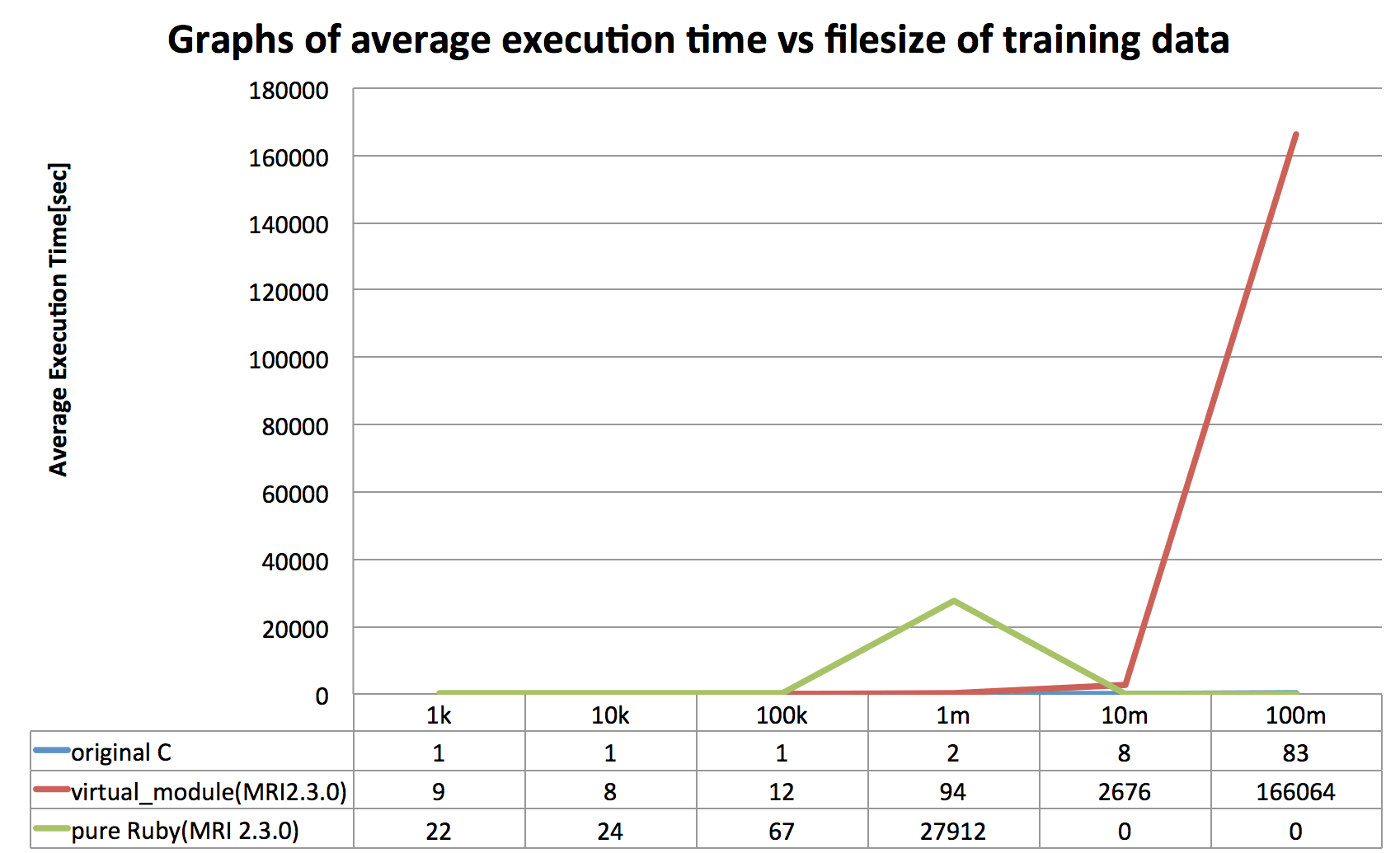
The other example here is a prototype word2vec implementation in Ruby, which is just partially ported (CBOW model only) from original C implementation (the one created by Tomas Mikolov at Google). Since it's not optimized very well yet due to my limited time to work on this example, it doesn't record decent score in terms of speed yet, but it reveals at least the fact that VirtualModule will reduces total execution time considerably even in real world example.
(A few record values resulted in 0 because it took too much time, actually more than three days to complete due to exponential explosion happened.)
Of course the vector binary file format generated through VirtualModule is compatible with original C implementation. Here is sample generated vector files saved onto GitHUb, and command logs are as shown below:
$ cd example
$ ruby word2vec.rb --output /tmp/vectors.bin --train ../doc/benchmark_word2vec/training_data/10mb.txt --size 20 --window 10 --negative 5 --sample 1e-4 --binary 1 --iter 3 --debug 0 > /dev/null 2>&1
$ python
Python 2.7.12 (default, Jul 1 2016, 15:12:24)
[GCC 5.4.0 20160609] on linux2
Type "help", "copyright", "credits" or "license" for more information.
>>> import gensim
>>> model = gensim.models.Word2Vec.load_word2vec_format('/tmp/vectors.bin', binary=True)
>>> model.most_similar("japan")
[(u'netherlands', 0.9741939902305603), (u'china', 0.9712631702423096), (u'county', 0.9686408042907715), (u'spaniards', 0.9669440388679504), (u'vienna', 0.9614173769950867), (u'abu', 0.9587018489837646), (u'korea', 0.9565504789352417), (u'canberra', 0.954473614692688), (u'erupts', 0.9540712833404541), (u'prefecture', 0.9534248113632202)]
To write Ruby code using VirtualModule is something like to write Python code using Cython. Basic concept is to separate large-scale computation algorithm, and isolate them as a module accessible from the base program. Like Cython, VirtualModule is NOT comprehensive approach too but gives you an opportunity to reduce execution time in exchange for the hard limitation of Ruby syntax due to ruby2julia transipiler.
require 'virtual_module'
vm = VirtualModule.new(methods:<<EOS)
def hi
"yo"
end
def init_table(list)
for i in 0..list.size-1
list[i]+=Random.rand
end
list
end
EOS
p vm.hi(:_) # "yo"
include vm
p init_table([1,20]) # [1.3066601775641218, 20.17001189249985]
As this sample snippet shows, VirtualModule is a Module generator which has only one public api named VirtualModule#new. Since VirtualModule#new returns a instance of Module class, user can simply include vm object that's why #init_table method is called in the context of self.
In detail, when you run VirtualModule, following processing are happening internally. It's just a typical procedures of an RPC call.
- VirtualModule calls
Julializer#ruby2juliato transpile#hiand#init_tablemethods into raw Julia code - Prepare glue code to send and receive parameters from/to Julia and add this to (1)
- Run (2) through Julia VM process and pass the parameters using msgpack-ruby and MsgPack.jl
- Receive returned value passed from Julia
What makes VirtualModule different from normal RPC call is #virtual_module_eval method. It offers a convenient way to eval arbitary code as if it's executed on the context of self. Most important note here is by this way, you can even pass type information about arguments like below.
# An example to pass Array{Float64,1} type array to Julia
class FloatArray < Array
end
float_list = FloatArray.new([1.0234, 9.9999])
vm.virtual_module_eval("float_list = init_table(float_list)")
puts float_list # [1.5765191145261321, 10.270808112990153] is calculated as Array{Float64,1} in Julia, which means much faster computation than Array{Any,1}!
- Mac OSX, Linux and Ruby (MRI) 2.1 and higher are supported.
- Either Julia or Docker installed on your machine is a must, although installing Julia is highly recommended for the performance reason.
Simply run the following command.
$ gem install virtual_module
$ julia -e 'Pkg.add("MsgPack"); Pkg.add("MsgPackRpcServer")'
Following features are implemented already:
- File-based IPC using msgpack-ruby and MsgPack.jl
- RPC-based IPC using msgpack-ruby and MsgPackRpcServer.jl
features to be implemented in the future (if needed/requested) are listed as follows:
- A bug fix for
#virtual_module_eval: return value has a bug whenever argument has more than two lines. - Improve julializer (transpiler) much better
- word2vec implementation is another interesting option to improve
- Other programming language support such as golang, ruby itself etc.
MIT