The f2PilotBE package is designed to calculate the similarity factor
ƒ2 based on pharmacokinetic profiles from pilot
bioavailability/bioequivalence (BA/BE) studies, as an alternative
approach to assess the potential of bioequivalence of a Test product in
comparison to a Reference product in terms of maximum observed
concentration (Cmax) and area under the concentration-time
curve (AUC).
This package is based on the publications of Henriques et al. (2023) [1,2], which propose the use of the geometric mean (Gmean) ƒ2 factor, for the comparison of the absorption rate (given by Cmax) of Test and Reference formulations. According to the articles, Gmean ƒ2 factor using a cut-off of 35 showed a good relationship between avoiding type I and type II errors [1,2].
The similarity ƒ2 factor is a mathematical index widely used to compare dissolution profiles, evaluating their similarity, using the percentage of drug dissolved per unit of time.
The similarity ƒ2 factor, proposed by Moore and Flanner in 1996 [3], is derived from the mean squared difference, and can be calculated as a function of the reciprocal of mean squared-root transformation of the sum of square differences at all points:
$$f_{2}=50\cdot\log\biggl(100\cdot\biggl[1+\frac{1}{n}\sum_{t=1}^{t=n}{(\bar{R}{t}-\bar{T}{t})^2} \biggr]^{-0.5}\biggr)$$
where
The ƒ2 similarity factor ranges from 0 (when
$\bar{R}{t}-\bar{T}{t}=100%$, at all
Figure 1 (from Henriques et al. (2023) [1]) presents the distribution of ƒ2 similarity factor as a function of mean difference. Form the ƒ2 equation, an average difference of 10%, 15%, and 20% from all measured time points results in a ƒ2 value of 50 (red dotted lines), 41 (green dotted lines) and 35 (blue dotted lines), respectively.
Figure 1. Distribution of ƒ2 similarity factor as a function of mean difference. ƒ2 similarity factor is derived from the mean squared difference and can be calculated as a function of the reciprocal of the mean squared-root transformation of the sum of square differences at all points. An average difference of 10%, 15%, and 20% from all measured time points results in a ƒ2 value of 50 (red dotted lines), 41 (green dotted lines) and 35 (blue dotted lines), respectively. From Henriques, S.C. et al. (2023). Pharmaceutics, 15(5), 1430 (DOI: 10.3390/pharmaceutics15051430).As proposed by Henriques et al. (2023), the concept of similarity factor ƒ2 can be applied as an alternative to the average bioequivalence analysis, for pilot BA/BE studies [1,2].
ƒ2 can be used to assess the similarity on the rate of drug absorption by normalizing Test and Reference mean concentration-time profiles to the maximum plasma concentration (Cmax) derived from the mean Reference profile, until Reference Cmax is observed (Reference tmax) [1,2] (Figure 2):
Figure 2. Estimation of ƒ2 similarity factor from mean concentration-time profiles.where
The similarity ƒ2 factor is calculated as
where
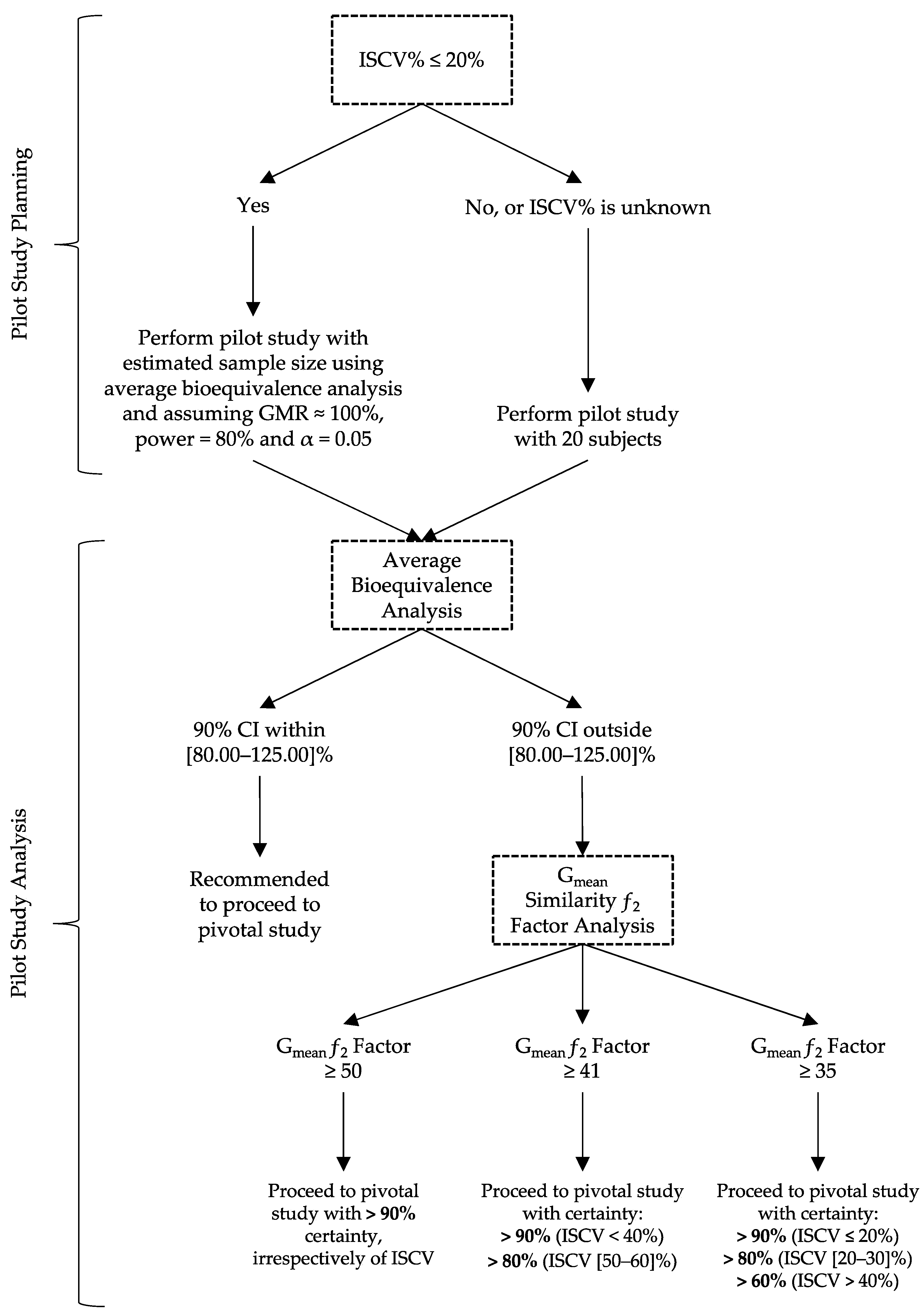
For the planning of pilot BA/BE studies, a decision tree is proposed (Figure 3, from Henriques et al. (2023) [2]).
For drug products with a known Intra-Subject Coefficient of Variation (ISCV%) below 20%, the authors propose the estimation of the sample size for a pilot study assuming a Test-to-Reference Geometric Least Square Mean Ratio (GMR) of 100%, a power of 80%, and an α of 0.05 [1,2]. However, for cases of higher ISCV% or unknown variability, it is propose the use of a fixed sample size of 20 subjects, as the use of higher sample sizes has not shown to increase the study power meaningfully, but was sufficient to avoid substantial type I errors [2].
Regarding the analysis of data from pilot studies, the authors propose to initially analyze the data using the average bioequivalence approach. For the case in which the calculated GMR and the corresponding 90% CI are not within [80.00–125.00]%, the alternative Gmean ƒ2 factor method should be used with a cut off of 35, as it was shown to be a valuable indicator of the potentiality of the Test formulation to be bioequivalent in terms of Cmax:
-
If the ƒ2 factor is above or equal to 35 (corresponding to a difference of 20% between Test and Reference concentration–time profiles until the Reference tmax), the confidence to proceed to a pivotal study is higher than 90% when ISCV% is lower or equal to 20%; the confidence is higher than 80% when ISCV% is within 20% and 30%; and the confidence is higher than 60% when ISCV% is higher than 40% [2].
-
If the ƒ2 factor is above or equal to 41 (corresponding to a difference of 15% between Test and Reference concentration–time profiles until the Reference tmax), the confidence to proceed to a pivotal study is higher than 90% for ISCV% until 40%, and higher than 80% for ISCV% within 50% to 60% [2].
-
If the ƒ2 factor is above or equal to 50 (corresponding to a difference of 10% between Test and Reference concentration–time profiles until the Reference tmax), the probability of the Test product to be truly bioequivalent to the Reference product in terms of Cmax, i.e., the confidence to proceed to a pivotal study, is higher than 90%, irrespective of the ISCV% [2].
The f2PilotBE is equipped with the following functions to aid in the
calculation of the similarity ƒ2 factor:
-
AUC()– calculates the cumulative area under the concentration-time curve (AUC) over time. -
AUClast()– calculates area under the concentration-time curve (AUC) from time zero to the time of the last measurable concentration (AUC0-t). -
Cmax()– calculates the maximum observed concentration post-dose (Cmax) directly obtained from the observed concentration-time profile. -
f2.AUC()– calculates the ƒ2 similarity factor for AUC, from concentration data. -
f2.Cmax()– calculates the ƒ2 similarity factor for Cmax, from concentration data. -
f2()– calculates the ƒ2 similarity factor, as proposed by Moore and Flanner in 1996. -
geomean()– calculates geometric mean, i.e., the Nth root of the product of the N observations, equivalent toexp(mean(log(x))). -
Tmax()– calculates the time of the Cmax.
To install the development version of f2PilotBE, start by installing
the devtools package from CRAN:
install.packages("devtools")Then the development version of f2PilotBE can be installed from GitHub
as:
library(devtools)
install_github("saracarolinahenriques/f2PilotBE")Following installation, load the f2PilotBE package and the ggplot2
package.
# Load packages
library("ggplot2") # required for plotting
library("f2PilotBE")As proposed by the authors [1,2], the ƒ2 similarity factor should be calculated from the arithmetic (Amean) or geometric (Gmean) concentration-time profiles.
The f2PilotBE provides a function (geomean()) for the calculation of
the Gmean. For the calculation of the Gmean
profile for Test and Reference products, first, import the individual
concentration-time data, and calculate the Gmean
concentration-time profiles for each Treatment, as follows.
# Import individual concentration data
dta_id <- read.csv('dta_id.csv')
# Calculate the geometric mean for each Treatment, by time point
dta <- data.frame(t(tapply(dta_id$Concentration,
dta_id[,c('Treatment','Time')],
geomean)))
dta$Time <- as.numeric(row.names(dta))
row.names(dta) <- c()The code above, will apply (tapply) the geomean function for each
given treatment, and each timepoint.
If the user prefers to use the Amean, instead of the
Gmean concentration-time data, simply replace the function
geomean, with the function mean from base R.
The f2.Cmax() function allows to calculate the ƒ2
similarity factor for Cmax, from Test and Reference mean
concentration-time profiles.
This function allows some flexibility on the structure of the mean
concentration-time dataframe. Treatment (Test and Reference) information
should either be provided in columns (pivoted) or rows (stacked). For
both cases a column with Time information should always be provided:
- For the case where Treatment information is provided in columns (pivoted), as in the following example:
# Create mean concentration-time data for Test and Reference product
# where treatment information is pivoted
dta_piv <- data.frame(
Time = c(0, 0.25, 0.5, 0.75, 1, 1.5, 1.75, 2, 2.25, 2.5,
2.75, 3, 3.25, 3.5, 3.75, 4, 6, 8, 12, 24),
Ref = c(0.00, 221.23, 377.19, 494.73, 555.74,
623.86, 615.45, 663.38, 660.29, 621.71,
650.33, 622.28, 626.72, 574.94, 610.51,
554.02, 409.14, 299.76, 162.85, 27.01),
Test = c(0.00, 149.24, 253.05, 354.49, 412.49,
530.07, 539.68, 566.30, 573.54, 598.33,
612.63, 567.48, 561.10, 564.47, 541.50,
536.92, 440.32, 338.78, 185.03, 31.13)
)
head(dta_piv)
#> Time Ref Test
#> 1 0.00 0.00 0.00
#> 2 0.25 221.23 149.24
#> 3 0.50 377.19 253.05
#> 4 0.75 494.73 354.49
#> 5 1.00 555.74 412.49
#> 6 1.50 623.86 530.07The ƒ2 similarity factor for Cmax can be
calculated from the mean concentration-time profiles, applying the
f2.Cmax() function as follows:
# Calculate f2 Factor for Cmax when treatment data is pivoted:
f2.Cmax(dta_piv, Time = "Time", Ref = "Ref", Test = "Test", plot = FALSE)
#> Cmax f2 Factor: 38.97As demonstrated above, for the case when treatment data is pivoted, the
f2.Cmax() function requires the indication of: (1) the dataframe with
mean concentration-time data (in this case dta = dta_piv), (2) the
name of the column in the dataframe with time information (in this case
Time = "Time"), (3) the name of the column in the dataframe with
concentration information for the Reference product (in this case
Ref = "Reference"), and (4) the name of the column in the dataframe
with concentration information for the Test product (in this case
Test = "Test").
Please note that Trt.cols is not required, as it defaults to TRUE
(i.e, logical value indicating whether treatment is presented in
columns/pivoted).
- For the case where Treatment information is provided in rows (stacked), as in the following example:
# Create mean concentration-time data for Test and Reference product
# where treatment information is stacked
dta_stk <- data.frame(
Time = rep(c(0, 0.25, 0.5, 0.75, 1, 1.5, 1.75, 2, 2.25, 2.5,
2.75, 3, 3.25, 3.5, 3.75, 4, 6, 8, 12, 24),2),
Trt = c(rep('R',20), rep('T',20)),
Conc = c(c(0.00, 221.23, 377.19, 494.73, 555.74,
623.86, 615.45, 663.38, 660.29, 621.71,
650.33, 622.28, 626.72, 574.94, 610.51,
554.02, 409.14, 299.76, 162.85, 27.01),
c(0.00, 149.24, 253.05, 354.49, 412.49,
530.07, 539.68, 566.30, 573.54, 598.33,
612.63, 567.48, 561.10, 564.47, 541.50,
536.92, 440.32, 338.78, 185.03, 31.13))
)
head(dta_stk)
#> Time Trt Conc
#> 1 0.00 R 0.00
#> 2 0.25 R 221.23
#> 3 0.50 R 377.19
#> 4 0.75 R 494.73
#> 5 1.00 R 555.74
#> 6 1.50 R 623.86tail(dta_stk)
#> Time Trt Conc
#> 35 3.75 T 541.50
#> 36 4.00 T 536.92
#> 37 6.00 T 440.32
#> 38 8.00 T 338.78
#> 39 12.00 T 185.03
#> 40 24.00 T 31.13The ƒ2 similarity factor for Cmax can be
calculated from the mean concentration-time profiles, applying the
f2.Cmax() function as follows:
# Calculate f2 Factor for Cmax when treatment data is stacked:
f2.Cmax(dta_stk, Time = "Time", Conc = "Conc",
Trt = "Trt", Ref = "R", Test = "T",
Trt.cols = FALSE, plot = FALSE)
#> Cmax f2 Factor: 38.97As demonstrated above, for the case when treatment data is stacked, the
f2.Cmax() function requires the indication of: (1) the dataframe with
mean concentration-time data (in this case dta = dta_stk), (2) the
name of the column in the dataframe with time information (in this case
Time = "Time"), (3) the name of the column in the dataframe with
concentration information (in this case Conc = "Conc"), (4) the name
of the column in the dataframe with treatment information (in this case
Trt = "Trt"), (5) the nomenclature in the dataframe for the Reference
product (in this case Ref = "R"), (6) the nomenclature in the
dataframe for the Test product (in this case Test = "T"), and (7)
indication that treatment is presented in rows/stacked
(Trt.cols = FALSE).
For both cases, a graphical representation of the normalized Test and
Reference concentrations over time, by Reference Cmax, until
Reference tmax, can be generated, by simply turning plot to
TRUE:
# Calculate f2 Factor for Cmax, and plot normalized concentration by Reference Cmax
f2.Cmax(dta_piv, Time = "Time", Ref = "Ref", Test = "Test", plot = TRUE)#> Cmax f2 Factor: 38.97
By default, f2.Cmax() plots the Reference product in black, and the
Test product in blue, nevertheless,
the user can personalize the output plot with different colours
according to their preference, by using col.R and col.T, for
Reference and Test product respectively:
# Calculate f2 Factor for Cmax, and plot normalized concentration by Reference Cmax
f2.Cmax(dta_piv, Time = "Time", Ref = "Ref", Test = "Test",
plot = TRUE, col.R = "darkblue", col.T = "red")#> Cmax f2 Factor: 38.97
-
Henriques, S.C.; Albuquerque, J.; Paixão, P.; Almeida, L.; Silva, N.E. (2023). Alternative Analysis Approaches for the Assessment of Pilot Bioavailability/Bioequivalence Studies. Pharmaceutics. 15(5), 1430. DOI: 10.3390/pharmaceutics15051430.
-
Henriques, S.C.; Paixão, P.; Almeida, L.; Silva, N.E. (2023). Predictive Potential of Cmax Bioequivalence in Pilot Bioavailability/Bioequivalence Studies, through the Alternative ƒ2 Similarity Factor Method. Pharmaceutics. 15(10), 2498. DOI: 10.3390/pharmaceutics15102498.
-
Moore, J.W.; Flanner, H.H. (1996). Mathematical Comparison of Curves with an Emphasis on in Vitro Dissolution Profiles. Pharm. Technol.. 20, 64–74.