Releases: simpeg/discretize
Pure Python 3D Viz Example
- from pr #135
- commits from: @banesullivan
- review from: @lheagy
This provides updates to the vtkModule documentation that reflect recent development in vtki and a new example for 3D visualization. Check out the new example and the types of 3D renderings that are now possible in a pure Python environment (also its Python 3 friendly!):
Example in Brief
Using vtki, any discretize mesh can now be immediately rendered using the toVTK() method. Be sure to check out the vtki docs to learn more about using vtki!
import discretize
import numpy as np
import vtki
vtki.set_plot_theme('document')
# Create a TensorMesh
h1 = np.linspace(.1, .5, 3)
h2 = np.linspace(.1, .5, 5)
h3 = np.linspace(.1, .8, 3)
mesh = discretize.TensorMesh([h1, h2, h3])
# Get a VTK data object
dataset = mesh.toVTK()
# Defined a rotated reference frame
mesh.axis_u = (1,-1,0)
mesh.axis_v = (-1,-1,0)
mesh.axis_w = (0,0,1)
# Check that the reference frame is valid
mesh._validate_orientation()
# Yield the rotated vtkStructuredGrid
dataset_r = mesh.toVTK()
p = vtki.BackgroundPlotter()
p.add_mesh(dataset, color='green', show_edges=True)
p.add_mesh(dataset_r, color='maroon', show_edges=True)
p.show_grid()
p.screenshot('vtk-rotated-example.png')Add vtki support to make using VTK data objects more Pythonic
- from pr #130
- commits from: @banesullivan
- review from: @lheagy
Add vtki support to make using VTK data objects more Pythonic
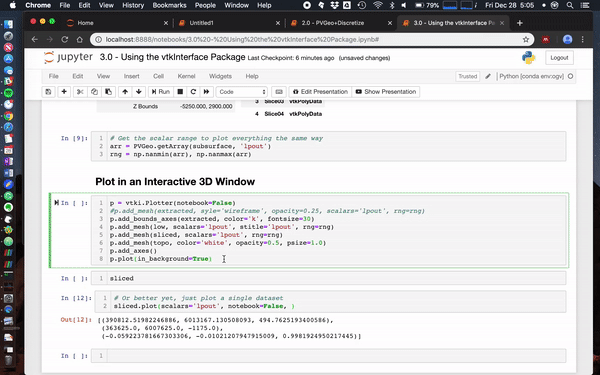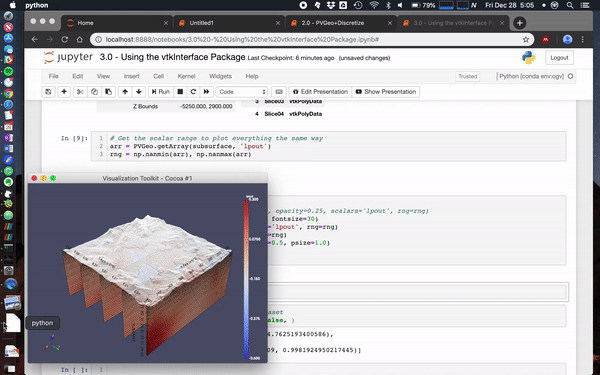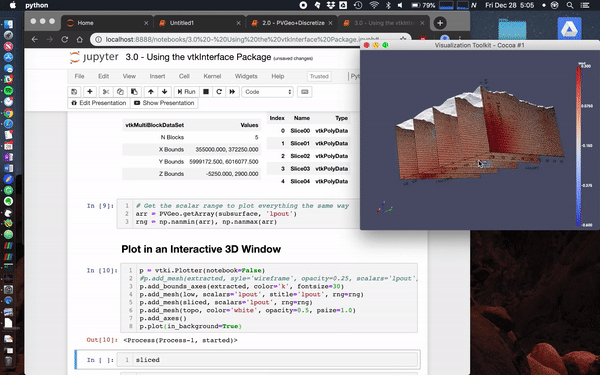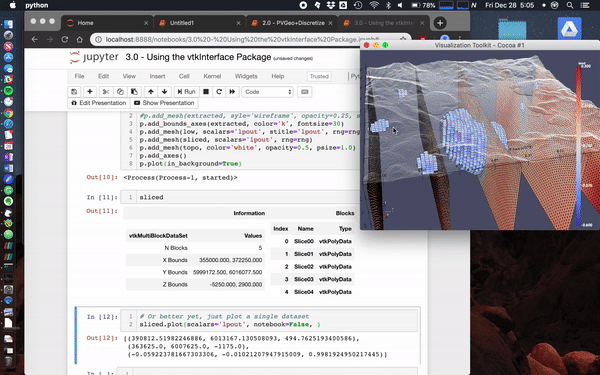
@banesullivan recently added a ton of new features to vtki (the VTK interface Python package) that help make using just about any VTK data objects more intuitive/Pythonic. If vtki is available in your Python environment then these changes make using VTK data objects way easier. Here's a screenshot of what this currently looks like in a Jupyter notebook (creates static renderings but can also be interactive in a separate pop-up window).
Also check out this notebook to see more ways to use PVGeo, discretize, and vtki. Below is a screenshot of a simple use case:
Simple example
import discretize
import numpy as np
# Create a simple TensorMesh
h1 = np.linspace(.1, .5, 3)
h2 = np.linspace(.1, .5, 5)
h3 = np.linspace(.1, .5, 3)
mesh = discretize.TensorMesh([h1, h2, h3])
# Convert to VTK object and use vtki to render it
mesh.toVTK().plot(notebook=False)Fancy example
Here we load the Laguna del Maule Bouguer Gravity example from the SimPEG docs.
This data scene was produced from the Laguna del Maule Bouguer Gravity example provided by Craig Miller (Maule volcanic field, Chile. Refer to Miller et al 2016 EPSL for full details.)
Miller, C. A., Williams-Jones, G., Fournier, D., & Witter, J. (2017). 3D gravity inversion and thermodynamic modelling reveal properties of shallow silicic magma reservoir beneath Laguna del Maule, Chile. Earth and Planetary Science Letters, 459, 14–27. https://doi.org/10.1016/j.epsl.2016.11.007
import shelve
import tarfile
import discretize
f = discretize.utils.download(
"https://storage.googleapis.com/simpeg/laguna_del_maule_slicer.tar.gz"
)
tar = tarfile.open(f, "r")
tar.extractall()
tar.close()
with shelve.open('./laguna_del_maule_slicer/laguna_del_maule-result') as db:
mesh = db['mesh']
Lpout = db['Lpout']
mesh = discretize.TensorMesh.copy(mesh)
models = {'Lpout':Lpout}
mesh.toVTK(models).plot()Usage Notes
Since vtk and vtki are not required dependencies you'll need to make sure you install them to your Python environment. Pip install for vtki should do the trick but Windows folks might need to install vtk from conda seperately. Also this is Python 3 friendly!
pip installl vtkiStream thickness
Description
Added stream_thickness keyword argument to plotSlice and _plotImage2D functions so that it is possible to scale the thickness of streamlines to reflect the amplitude of the vector field. This functionality was added in a manner similar to the stream_threshold keyword.
stream_thickness keyword currently takes a float which acts as a scaling factor for the streamline thickness. Bounds are hardwired to fix the thickness of the 10% largest and smallest vector amplitudes. Provides good results with the DC current density plots I've made but could probably be generalized in the future for more flexibility.
e.g.
mesh.plotSlice(
u, ax=ax, normal='X', vType='F', view='vec', stream_threshold=1e-8, stream_thickness=3
)Expand the `plot_3d_slicer` to other `vTypes` and `views`
Expand plot_3d_slicer
Addresses #116 .
Bug fix
First, it contains a minor bug-fix in the scrolling behavior (last element in each direction was not acceptable, only noticeable in small grids).
vType
Included all non-vector vType's: CC, Ex, Ey, Ez, Fx, Fy, Fz.
view
Included all view's except vec (real [default]; imag; abs; tested them all, seems to be fine).
Name-clash
There is a problem with the view-parameter, which I stupidly used to switch the x-y-axis. I changed the previous view-parameter to axis (hence axis='xy' or 'yx'). It is better to change this than to have different parameters as, for instance, in plotSlice. I added a switch for backwards-compatibility (if view in ['xy', 'yx'] => it sets axis = view; view = 'real').
Add VTK object interface
- from pr: #114
- commits from: @banesullivan
- review from: @rowanc1, @lheagy
Description
These new features enable discretize to have a direct interface for VTK base software by implementing toVTK() methods on all the mesh types (excluding CylMesh at this time). Notably, @banesullivan will be using this to provide interoperability with PVGeo to provide direct file IO into ParaView using discretize as well as ways to interactively create discretize meshes in ParaView similar to this example in the PVGeo docs. This new interface also enables discretize meshes to be passed on directly to VTK algorithms for post-processing analysis (note if you couple the interface with PVGeo like shown in this notebook, the process is somewhat simplified).
To learn more about the new VTK object interface, see the docs for the vtkInterface.
Example Usage
The VTK object interface can be used on TensorMesh, TreeMesh, and CurvilinearMesh objects to yield a VTK object in your active Python environment or used to save VTK files for easy sharing.
import discretize
import numpy as np
h1 = np.linspace(.1, .5, 3)
h2 = np.linspace(.1, .5, 5)
h3 = np.linspace(.1, .5, 3)
mesh = discretize.TensorMesh([h1, h2, h3])
# Yield a VTK data object for passing this mesh onto VTK algorithms
vtkobj = mesh.toVTK()
# Or save out the tree mesh for sharing with others
mesh.writeVTK('sample_mesh')Note that these new features also give users the ability to specify the axes orientation of any given mesh. For example, the above TensorMesh is oriented on the traditional Cartesian reference frame by default but we could change this. To define what that orientation is, we can use the new axis_* properties:
# Defined a rotated reference frame
mesh.axis_u = (1,-1,0)
mesh.axis_v = (-1,-1,0)
mesh.axis_w = (0,0,1)
# Check that the reference frame is valid
mesh._validate_orientation()Now we have a TensorMesh explicitly defined on a rotated reference frame! This is quite useful for when we want to convert this to a VTK data object that must have its location in a traditional XYZ Cartesian space defined.
Please take a look at the docs to learn more about using these new features!
plot_3d_slicer
plot_3d_slicer
Add an interactive slicer for 3D volumes. At the moment only implemented for tensor meshes.
Features
- Mouse wheel scroll while hovering over a subplot scrolls through the third axis (e.g., hovering over the xy-slice and scrolling your mouse wheel will go through the z-axis).
- The three subplots are synced, also for zooming and moving.
- The initial slices can be provided via the
xslice,yslice, andzsliceparameters (default is in the middle of the volume). - Transparency values and ranges can be provided (a list of floats and tuples/lists of two values), e.g. to hide the seawater or to focus on an interesting part, e.g.,
[0.3, [1, 4], [-np.infty, -10]]to remove all values equal to 0.3, all values between 1 and 4, and all values smaller than -10. For interactive range selection settransparency='slider'. - Takes
climandpcolorOptsas other mesh-plotting functions, which will be passed topcolormesh. - By default the horizontal axis is
x, and the vertical axis isy; this can be flipped by settingview='yx'.
By default, the aspect ratio of the three subplots is set to 'auto'. You can change this with the aspect
parameter, however, expect the unexpected by doing this. Most importantly, the three subplots won't be nicely aligned, and zooming might result in funny arrangements. Two parameters can be used in this respect:
aspecttakes'auto','equal', ornum. A list of two of them can be provided, in which case the first element is for the xy-slice, and the second element for the xz- and zy-slices. E.g.,aspect=['equal', 2]sets the xy-slice toequal, and in the other two the vertical dimension is exaggerated by a factor of 2.- The
plot_3d_sliceris on asubplot2grid-grid, by default on a 3x3 grid, where 2x2 are used for the xy-slice, 2x1 for the xz-slice, and 1x2 for the zy-slice. You can provide a list of three integers via thegrid-parameter, which stand for the number of grid-units occupied for the x-, y-, and z-dimension (default is[2, 2, 1]).
Usage
mesh.plot_3d_slicer(data)
It requires %matplotlib notebook in Jupyter. In regular IPython shells it should just work.
options on plotSlice, grid=True
Include tree.pxd in source distribution
Minor update: plotGrid - linestyle
- from pr #101
".-"is no longer valid for a linestyle input in matplotlib, it should instead be "-." (however, a solid line looks better anyways for the 1d).
TreeMesh Re-write
New Implementation of the TreeMesh
At this point consider this branch as EXPERIMENTAL. There are many possible unsafe operations that could arise, so be careful (which will need to be enforced at a later time). The code is certainly not completely up to format standards, but at this point, just wanted to get a pull request going and allow anyone who wants to use it to test it out to find any bugs.
There are many changes to the TreeMesh implementation within this pull request, However it is mostly feature complete compared to the previous implementation with regards to the public members of the class.
It was mostly rewritten in a way that made the construction of the mesh and construction of the operators all done in c++/cython, which resulted in dramatic speedups. As a reference, the 97 nosetests on the tree in the previous implementation took 238.476s on my personal computer. This implementation took 20.281s.
The basic idea is that each object, (node, edge, etc.) is aware of the structure of the TreeMesh as each is a cpp class that contains references to other objects.
A few key differences:
-
All tree construction is balanced (no need to call
tree.balance, or passbalance=Trueto functions) -
tree.refineshould only be called once (at this point) as it "finalizes" the tree. It might be good to add a flag to the tree initialization that would allow incremental additions (similar toscipy.spatial.ConvexHull) and then require a finalization operation to be done before any other operations. -
Interpolation is "lazy" 2nd order now for all
E,CC, andFinterpolation, meaning we triangulate the grid points to interpolate. Interpolating without triangulation on the TreeMesh is still a point of research. This is not as fast as it could be, but still faster than previously. -
Both
faceandedgeoperations are defined for 2D, (basically a re-ording of x-edges -> y-faces) -
Do not expect any ordering for the faces, edges, and nodes. They are whatever they have decided to be.
-
Do not expect any of the private members of the class to have remained consistent between implementations.
-
There are many other changes which should hopefully be apparent as you look through the code.
Big things that still need to be implemented
-
Serialization, (do not expect to pickle this object and have it work, however with construction being much faster, this shouldn't slow down anyone too much while it is being worked on).
-
PlotImageandPlotSlice.
There are also many areas that this code could be extended to handle different types of underlying meshs, as well as support for differing sizes of axes, but as I said before, the initial goal of this pull request is to mimic the behavior of the current implementation.
Other than that, I hope this speeds up the operations for those who need them.
Update
- Serialization should be implemented now (the
TreeMeshis pickle-able) - PlotImage and PlotSlice are also implemented.
- It also now support differing dimensions of the underlying
TensorMesh - Interpolation is now NOT triangulated, so do there is no longer 2nd order convergence of the non-node interpolation matrices.
- the
refineandinsert_cellsfunction now have afinalizekeyword arg that can be set to false if you want to do multiple steps of tree building