-
Notifications
You must be signed in to change notification settings - Fork 0
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Direct Fourier transform prediction errors #34
Comments
|
Hi Ben,
Thanks I will have a look at all of this and try to track down the problem.
It would be great if you could send along your notebook and possibly point
me to the ms you are using so we can be sure I am using the exact same data
for verification purposes.
Cheers,
Kim
…On Mon, Sep 7, 2020 at 11:45 AM Benjamin Hugo ***@***.***> wrote:
Hi @KimMcAlpine <https://github.com/KimMcAlpine>, @ludwigschwardt
<https://github.com/ludwigschwardt>
I have done some preliminary comparisons with RARG's crystalball which is
used in production continuum subtraction and has been verified against the
Meqtrees simulator. I used your notebook invoking your DFT implementation
to simulate model data and repack it into a measurement set column for
storage and comparison. Notebook is available on request, but I will list
the code below for reference discussion. The two simulations must match to
machine precision, because they both use the SIN projection and UVW
coordinate frame.
This simple comparison was needed after I spotted that the residual
product formed between my CORRECTED_DATA and your limited model was offset
and therefore not subtracting properly:
[image: image]
<https://user-images.githubusercontent.com/7934586/92372800-bdcde180-f0fd-11ea-92e9-81b801bb911c.png>
To verify that this is not a model error I simplified the comparison to a
single component. The two models I used are one to one matches with a
single point source at the position of 1934, slightly off the pointing
center (I think the 2019 September OPT coordinate frames still slightly off
?!) of an actual UHF observation.
# References:
# Flux model use wsclean format
# Fluxes and positions from B.Hugo model in Modelling of PKS B1934-638 (J193925.0-634245) as UHF and L-Band calibrator for MeerKAT
#
fake, radec, 19:39:25.02671, -63.42.45.6255, 544 1088 wsclean 5.0 [0 0 0 0 0] false 815867187.5
vs
Format = Name, Type, Ra, Dec, I, SpectralIndex, LogarithmicSI, ReferenceFrequency='815867187.5', MajorAxis, MinorAxis, Orientation
fake,POINT,19:39:25.02671,-63.42.45.6255,5.0,[0.0,0.0,0.0,0.0,0.0],False,815867187.5,,,
I then compare a number of samples from predicted visibilities. Something
is clearly off int eh SP DFT (channel 1024 of 4096 and baseline m000&m005,
flags from observed visibilities applied to both sets):
[image: image]
<https://user-images.githubusercontent.com/7934586/92370323-6a0dc900-f0fa-11ea-8ee1-a5b92d98e6ec.png>
[image: image]
<https://user-images.githubusercontent.com/7934586/92370361-7a25a880-f0fa-11ea-98a0-bab01df9207b.png>
So as far as I can tell your DFT implementation is wrong even in the
narrow field case and we will have to track down where the bug is within
the software. The issue is, as far as I can tell not the models, although I
should add that you should collapse the components of the central source
into a single component for maximum precision as I requested per email
initially, but that is a different issue. I can do that for you and upload
it as another PR.
The notebook sections I list below for reference:
import katdal
import matplotlib.pyplot as plt
import numpy as np
import katpoint
from numba import jit, guvectorize
from katsdpcal.calprocs import get_bls_lookup, FluxDensityModel, K_ant, add_model_vis
from katsdpcal.calprocs_dask import wavg_full, bp_fit
from katsdpcal import pipelineprocs
import katsdpcal
import time
import dask.array as da
import glob
import os
from pyrap.tables import table as tbl
from pyrap.tables import taql
from pyrap.quanta import quantity
from datetime import datetime
f = katdal.open('1569150359_sdp_l0.full.rdb')
MSNAME = "1569150359_1934_XXYY_corrected_scan1.ms"
ms = tbl(MSNAME,readonly=False)
msant = tbl("{}::ANTENNA".format(MSNAME))
msspw = tbl("{}::SPECTRAL_WINDOW".format(MSNAME))
mspol = tbl("{}::POLARIZATION".format(MSNAME))
f.select()
print("Available targets:")
for t in f.catalogue.targets:
print("\t {}".format(t.name))
print("Available tracking scans:")
tracking_scans = list(filter(lambda x: x[1] == "track", list(f.scans())))
for t in f.catalogue.targets:
print("\t{}: ".format(t.name) +
", ".join([str(s[0]) for s in filter(lambda x: x[2].name == t.name, tracking_scans)]))
fieldsel = "J1939-6342"
onlyscans = [1]#None # None for all
scansel = list(filter(lambda x: x[2].name == fieldsel, tracking_scans))
#MODEL_DIRECTORY = os.path.join(katsdpcal.__path__[0], "conf", "sky_models")
MODEL_DIRECTORY = "."
print("Available models:")
for m in glob.glob(os.path.join(MODEL_DIRECTORY,"*.txt")):
print("\t{}".format(m))
# get model parameters, model params is a list of katpoint targets, or slightly modified katpoint targets
target = list(filter(lambda x: x.name == fieldsel, f.catalogue.targets))[0]
# The looks for a file which begins with the target name in the directory given in the 2nd arguement
model_params, model_file = pipelineprocs.get_model(target, MODEL_DIRECTORY, 'u')
print("Using model file '{}'".format(model_file))
ms_scans = list(sorted(np.unique(ms.getcol("SCAN_NUMBER"))))
assert len(ms_scans) == len(scansel), "Number of scans in katdal database and ms must match"
katdal2ms_scan_map = dict(zip([s[0] for s in scansel], ms_scans))
def baseline_index(a1, a2, no_antennae):
"""
Computes unique index of a baseline given antenna 1 and antenna 2
(zero indexed) as input. The arrays may or may not contain
auto-correlations.
There is a quadratic series expression relating a1 and a2
to a unique baseline index(can be found by the double difference
method)
Let slow_varying_index be S = min(a1, a2). The goal is to find
the number of fast varying terms. As the slow
varying terms increase these get fewer and fewer, because
we only consider unique baselines and not the conjugate
baselines)
B = (-S ^ 2 + 2 * S * # Ant + S) / 2 + diff between the
slowest and fastest varying antenna
:param a1: array of ANTENNA_1 ids
:param a2: array of ANTENNA_2 ids
:param no_antennae: number of antennae in the array
:return: array of baseline ids
"""
if a1.shape != a2.shape:
raise ValueError("a1 and a2 must have the same shape!")
slow_index = np.min(np.array([a1, a2]), axis=0)
return (slow_index * (-slow_index + (2 * no_antennae + 1))) // 2 + \
np.abs(a1 - a2)
def ants_from_baseline(bl, nant):
"""
Given unique baseline index and number of antennae lookup the corresponding
antenna indicies (zero-indexed).
:param bl: unique baseline index
:param nant: number of antennae
:return: pair of antenna indicies corresponding to unique index
"""
lmat = np.triu((np.cumsum(np.arange(nant)[None, :] >= np.arange(nant)[:, None]) - 1).reshape([nant, nant]))
v = np.argwhere(lmat == bl)
return v[0,0], v[0,1]
antennas = f.ants
antenna_names = [a.name for a in antennas]
array_position = katpoint.Antenna('', *antennas[0].ref_position_wgs84)
channel_freqs = f.freqs
antnames_ms = msant.getcol("NAME")
assert len(antenna_names) == len(antnames_ms) and antenna_names == antnames_ms, "katdal and ms antennas must match in ordering"
assert all(np.abs(msspw.getcol("CHAN_FREQ").ravel() - channel_freqs) < 1.0e-5), "katdal and ms channel ranges must match"
assert list(mspol.getcol("CORR_TYPE").ravel()) == [9,10,11,12], "MS must only have products XX, XY, YX, YY"
f.select()
for s in onlyscans if onlyscans is not None else [s[0] for s in scansel]:
f.select(targets=fieldsel, scans=[s], pol='h')
mssel = taql("select * from $ms where SCAN_NUMBER == {}".format(katdal2ms_scan_map[s]))
timestamps = f.timestamps
kd_delta_t = timestamps[1:] - timestamps[:-1]
assert all(kd_delta_t > 0) and all(kd_delta_t - kd_delta_t[0] < 1e-4), \
"Monotonically increasing katdal database needed"
print("Verifying ms vs. katdal indexing for scan {} (ms scan {})... ".format(s, katdal2ms_scan_map[s]), end="")
a1_ms = mssel.getcol("ANTENNA1")
a2_ms = mssel.getcol("ANTENNA2")
timestamps_ms = mssel.getcol("TIME")
uvw_ms = mssel.getcol("UVW")
flag_ms = mssel.getcol("FLAG")
blindx_ms = baseline_index(a1_ms, a2_ms, len(antnames_ms))
for bli in np.unique(blindx_ms):
(bla1, bla2) = ants_from_baseline(bli, len(antnames_ms))
blsel = np.logical_or(np.logical_and(a1_ms == bla1, a2_ms == bla2),
np.logical_and(a2_ms == bla1, a1_ms == bla2))
ms_delta_t = timestamps_ms[blsel][1:] - timestamps_ms[blsel][:-1]
assert all(ms_delta_t > 0) and all(ms_delta_t - ms_delta_t[0] < 1e-4), \
"Monotonically increasing measurement set database needed"
assert ms_delta_t[0] - kd_delta_t[0] < 1e-4, "katdal sampling rate must match ms sampling rate"
mjd_0 = quantity("{}s".format(timestamps_ms[blsel][0])).to_unix_time()
# starting time of scan must match that of ms, monotonically increasing at same rate
assert np.abs(mjd_0 - timestamps[0]) < 1e-4, "katdal timestamps must match ms timestamps"
assert timestamps.size == timestamps_ms[blsel].size, "katdal timestamps must match ms timestamps"
print("<done>")
print("Predicting scan {} (ms scan {})... ".format(s, katdal2ms_scan_map[s]), end="")
bls_lookup = get_bls_lookup(antenna_names, f.corr_products)
uvw = target.uvw(antennas, timestamps, array_position)
uvw = np.array(uvw, np.float32)
# set up model visibility
ntimes, nchans, nbls = f.vis.shape
nants = len(f.ants)
assert a1_ms.size == ntimes * nbls, "Number of baselines in ms and katdal dataset does not match"
# currently model is the same for both polarisations
# TODO: include polarisation in models
k_ant = np.zeros((ntimes, nchans, nants), np.complex64)
complexmodel = np.zeros((ntimes, nchans, nbls), np.complex64)
complexmodel_ms = np.zeros((a1_ms.shape[0], nchans, 4), np.complex64)
assert nbls == bls_lookup.shape[0]
wl = katpoint.lightspeed / f.channel_freqs
for ss in model_params:
model_targets = katpoint.Target(','.join(ss.split(',')[:-1]))
model_fluxes = FluxDensityModel(''.join(ss.split(',')[-1]))
S=model_fluxes.flux_density(f.channel_freqs/1e6)
l, m = target.sphere_to_plane(
*model_targets.radec(), projection_type='SIN', coord_system='radec')
k_ant = K_ant(uvw, l, m, wl, k_ant)
complexmodel = add_model_vis(k_ant, bls_lookup[:, 0], bls_lookup[:, 1],
S.astype(np.float32), complexmodel)
for kda1, kda2 in np.unique(bls_lookup, axis=0):
kdblsel = np.logical_or(np.logical_and(bls_lookup[:,0] == kda1,
bls_lookup[:,1] == kda2),
np.logical_and(bls_lookup[:,0] == kda2,
bls_lookup[:,1] == kda1))
msblsel = np.logical_or(np.logical_and(a1_ms == kda1,
a2_ms == kda2),
np.logical_and(a1_ms == kda2,
a2_ms == kda1))
# select all times of this baseline into row format
# both correlations the same, ie. I +/- Q:=0
assert np.sum(kdblsel) == 1, "debug: one baseline must be selected"
complexmodel_ms[msblsel,:,0] = complexmodel[:, :, kdblsel].reshape(ntimes, nchans)[:, :]
complexmodel_ms[msblsel,:,3] = complexmodel[:, :, kdblsel].reshape(ntimes, nchans)[:, :]
print("<done>")
print("Writing model for ms scan {}... ".format(katdal2ms_scan_map[s]), end="")
mssel.putcol("MODEL_DATA", complexmodel_ms)
print("<done>")
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#34>, or unsubscribe
<https://github.com/notifications/unsubscribe-auth/ACMD2WJSXCLIXGAIFSXLOOLSESTSFANCNFSM4Q53LZYA>
.
|
|
Hi Kim: |
|
@marisageyer @sbuchner as discussed until this issue is resolved and the DFT is properly tested I highly recommend that the codepath be disabled in phaseups as it will induce decoherency by artificially shifting the beam around on both L and UHF band beamformer phaseup and cause,by the looks of it, astrometrical errors on calibrated imaging data produced by the SP pipeline. @ludwigschwardt is there a way to quickly kill this code path until resolution? |
|
@KimMcAlpine would know best - maybe go back to the old single component model which doesn't need a DFT? |
|
I think to achieve this we just need to replace the model files in the conf
directory with ones that contain only a single source. So it should be
fairly straightforward to achieve.
…On Tue, Sep 8, 2020 at 10:11 AM Ludwig Schwardt ***@***.***> wrote:
@KimMcAlpine <https://github.com/KimMcAlpine> would know best - maybe go
back to the old single component model which doesn't need a DFT?
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#34 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/ACMD2WISVFLO3UBP63732Y3SEXRL7ANCNFSM4Q53LZYA>
.
|
|
Hi @ludwigschwardt and @KimMcAlpine - quick check: while the multi-component models were introduced at the end of June (28 June), do we know whether this DFT bug that @bennahugo describes necessarily would have been introduced at the same time? Or could it be that even pre the update multi-component models such a bug (leading to an offset between model and source) exists? -- answering myself: from above comments its clear the single comp models don't need a DFT. |
|
@bennahugo I think I have discovered the reason for the discrepancy in the plots above. It was a bit confusing initially because I thought you were plotting samples for one baseline, but I suspect that maybe you are plotting a subsample of the baselines at some timestamps. I have compared my SP model with the 'corrected data column' in the indicated MS I think the problem is with your model string which you provide as being: If I use this string to form the complex model and compare the SP predicted model and the corrected_data column in the MS for baseline m000-m005 at channel 1024 for the first scan I get the following plot: If I plot a subsample of baselines at timestamp 10 I get: If I modify the string to be: As I'm not sure exactly which samples you plotted before I can't compare directly, but this looks like its the most plausible reason for the discrepancy in the latest test. The comparison between SDP and wsclean looks good to be here to me in the narrow field. Let me know what kind of tests you want to proceed with from here. We obviously still need to deal with the phase offset from your earlier test. |
|
so this is just a question of ":" in stead of "." in the declination? |
|
Yes, I think so :)
…On Thu, Sep 10, 2020 at 2:22 PM Tony Foley ***@***.***> wrote:
so this is just a question of ":" in stead of "." in the declination?
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#34 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/ACMD2WINOFOJ6TRXONFQQLDSFDAJRANCNFSM4Q53LZYA>
.
|
|
Okay thanks @KimMcAlpine. You are correct I forgot to apply the selection mentioned. When I changed the coordinates as mentioned and increased the offset to about a degree from phase centre I notice larger offsets between the two implementations on the long spacings: Now I think the errors on the long spacings are too big to safely ignore - the error in the absolute anglular difference is nearly 3 arcsecs at the 75th percentile, while the power is almost spot on. I think this is responsible for the subtraction differences. So there is a systemic error in the phase which could stem from the uv coordinates (which it should not - both are ultimately generated by katpoint) or an error in the wavelengths applied. Are we sure that the cosines are computed in double precision? |
|
Thanks Ben, can you point me to the MS you used and send along the
co-ordinates you've used in the test so I can track down the problem.
…On Mon, Sep 14, 2020 at 11:16 AM Benjamin Hugo ***@***.***> wrote:
Okay thanks @KimMcAlpine <https://github.com/KimMcAlpine>. You are
correct I forgot to apply the selection mentioned. When I changed the
coordinates as mentioned and increased the offset to about a degree from
phase centre I notice larger offsets between the two implementations on the
long spacings:
[image: image]
<https://user-images.githubusercontent.com/7934586/93065767-6ab5da80-f679-11ea-8c09-1b03e6d5d5ba.png>
compared to the short spacings whic should be ok
[image: image]
<https://user-images.githubusercontent.com/7934586/93065971-a8b2fe80-f679-11ea-81c5-38f7ed53f4e1.png>
Now I think the errors on the long spacings are too big to safely ignore -
the error in the absolute anglular difference is nearly 3 arcsecs, while
the power is almost spot on. I think this is responsible for the
subtraction differences. So there is a systemic error in the phase which
could stem from the uv coordinates (which it should not - both are
ultimately generated by katpoint) or an error in the wavelengths applied.
Are we sure that the cosines are computed in double precision?
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#34 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/ACMD2WOAZXOR5FN4LAMLCJLSFXNOBANCNFSM4Q53LZYA>
.
|
|
Same MS, with an offset position to the south of the phase centre |
|
Could you copy the MS? I'm trying to further check the dtypes being used and the coordinates against that generated by astropy |
|
@KimMcAlpine I believe I've tracked down the issue to some extent. There is a difference between the uvw coordinates you get out of katpoint and the ones that are in the MS via katdal. Now the question is what are you doing in the sp pipeline @ludwigschwardt ? |
|
Yea that was the bastard @KimMcAlpine. Can you please verify that this is done in full precision in the actual pipeline? |
Interesting... Because the precision is 64-bit at the start. In [57]: f = katdal.open('.....rdb')
In [58]: target = f.catalogue.targets[0]
In [59]: uvw = target.uvw(f.ants, f.timestamps, f.ants[0].array_reference_antenna())
In [60]: uvw[0].dtype
Out[60]: dtype('float64')I'll dig out the relevant dataset (1569150359) and check katdal vs katpoint UVWs, first leaving MS out of the loop to reduce the number of factors to consider. |
|
In the pipeline I'm using 32-bit precision. I probably did this to speed things up a bit, the code in the pipeline is given below: |
|
Aside from memory allocation and setting, speed should not be an issue here (at least not on CPUs). As mentioned this causes arcsecond level errors and should be changed. |
|
@bennahugo, in the Python example you posted above, how did you generate the |
|
That was cast to np.float32 in the array @ludwigschwardt as @KimMcAlpine has above. |
|
Right, that makes more sense now. The difference is 0.0002 metres, or 0.2 mm, though. On an 8000 m baseline this amounts to 5 milliarcseconds, a few orders below the 3 arcsecond position errors you mention. |
|
@ludwigschwardt this is a degree away at phase centre (at delta_ra=0). According to my calculation the phase differences, at the 75th percentile is around the 2" mark so it is not far off the level of errors that we are seeing if you look at the first plot |
|
You mention a 2-3 arcsecond phase difference. Is this a difference in the phase angle of the calibrated visibilities of the calibrator as produced by two different methods, or a position offset on the sky? If the former, it would be informative to express this error as a noise RMS, especially since your plots suggest that the mean phase residual is zero and the phase difference is in fact the maximum deviation of the residual noise. If mostly due to float32 usage we can think of this as quantisation noise. We can then compare this to the expected noise level to see if it is excessive. If the latter, I'd be more surprised. There is a known issue with katpoint astrometric accuracy at the 2 arcsecond level, but this would manifest itself quite far away from the calibrator (30 degrees+). |
|
it is the phase angle difference of your predicted model visibilities vs our predicted model visibilities on a source placed at an offset position @ludwigschwardt. The error is solely due to quantization - without casting down the error on short and long spacings is far more comparable than when the decomposed uvw coordinates are quantized before combining into a baseline uvw pair. I think the problem with exposing this as noise RMS is that it becomes sky dependent: dependent on how bright the source is and how far off axis it is. Surely changing the dtype here will have no effect aside from avoiding running back into this issue as we extend the baselines even further. I would understand the argument from a performance point of view if we were talking about a cuda / opencl based dft, but this is not the case here? |
|
I agree that there is no real need for 32-bit precision and we should go back to full precision. I'm just curious to understand the size of this effect. As I see it this will cause a slight increase in the noise floor, but how slight is the question :-) |
|
@KimMcAlpine are you planning to implement this change in precision? Thereafter @bennahugo can check that the phase offsets between the two sets of tools look good on both long and short baselines. |
|
Yes, I'll have a PR later today.
…On Thu, Sep 17, 2020 at 9:08 AM marisageyer ***@***.***> wrote:
@KimMcAlpine <https://github.com/KimMcAlpine> are you planning to
implement this change in precision? Thereafter @bennahugo
<https://github.com/bennahugo> can check that the phase offsets between
the two sets of tools look good on both long and short baselines.
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#34 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/ACMD2WIZA74F32BRSKKQSADSGGYXLANCNFSM4Q53LZYA>
.
|
During the model prediction the uvw co-ordinates were being cast to np.float32 precision which causes phase errors on sources situation some distance from the phase centre as discussed in #34. This PR restores the uvw co-ordinates to their native np.float64 precision.











Hi @KimMcAlpine, @ludwigschwardt
I have done some preliminary comparisons with RARG's crystalball which is used in production continuum subtraction and has been verified against the Meqtrees simulator. I used your notebook invoking your DFT implementation to simulate model data and repack it into a measurement set column for storage and comparison. Notebook is available on request, but I will list the code below for reference discussion. The two simulations must match to machine precision, because they both use the SIN projection and UVW coordinate frame.
This simple comparison was needed after I spotted that the residual product formed between my CORRECTED_DATA and your limited model was offset and therefore not subtracting properly:
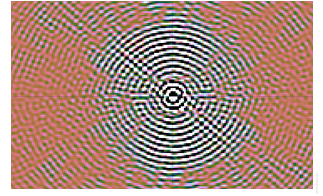
The minimum and maximum -7.2 and +6.9 Jy
To verify that this is not a model error I simplified the comparison to a single component. The two models I used are one to one matches with a single point source at the position of 1934, slightly off the pointing center (I think the 2019 September OPT coordinate frames still slightly off ?!) of an actual UHF observation.
vs
I then compare a number of samples from predicted visibilities. Something is clearly off int eh SP DFT (channel 1024 of 4096 and baseline m000&m005, flags from observed visibilities applied to both sets):
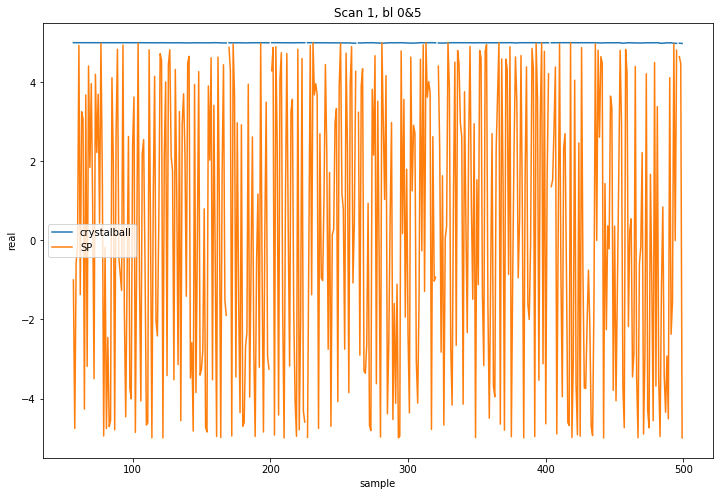
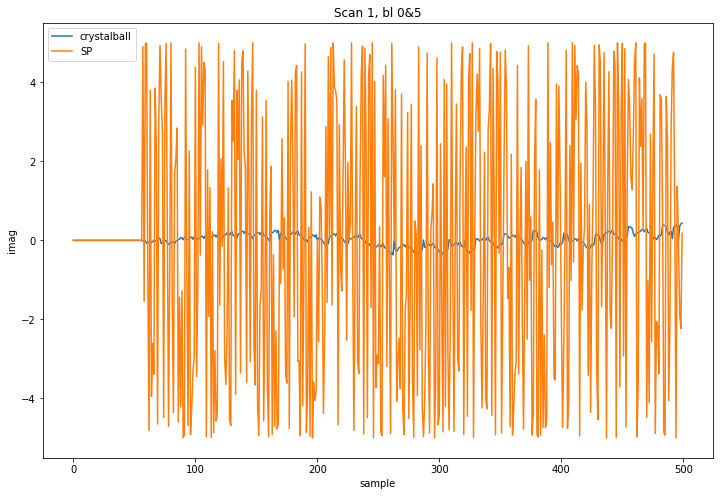
So as far as I can tell your DFT implementation is wrong even in the narrow field case and we will have to track down where the bug is within the software. The issue is, as far as I can tell not the models, although I should add that you should collapse the components of the central source into a single component for maximum precision as I requested per email initially, but that is a different issue. I can do that for you and upload it as another PR.
The notebook sections I list below for reference:
The text was updated successfully, but these errors were encountered: