-
Notifications
You must be signed in to change notification settings - Fork 10
What is Inside a BioGraph
A BioGraph consists of a directory ending in .bg with several files beneath it.
$ find HG002.bg/ -type f
HG002.bg/analysis/results.vcf.gz
HG002.bg/analysis/results.vcf.gz.tbi
HG002.bg/coverage/51b8861451bcccc1c2f8e5cc76233f6ba6e801fd.readmap
HG002.bg/metadata/bg_info.json
HG002.bg/qc/classifier_log.txt
HG002.bg/qc/create_log.txt
HG002.bg/qc/create_stats.json
HG002.bg/qc/kmer_quality_report-BELOW_MIN_COUNT.html
HG002.bg/qc/kmer_quality_report.html
HG002.bg/qc/timings.json
HG002.bg/qc/variants_log.txt
HG002.bg/qc/variants_stats.json
HG002.bg/seqsetMost of the data is kept in the seqset file and the coverage directory:
$ du -sch HG002.bg/*
294M HG002.bg/analysis
7.0G HG002.bg/coverage
8.0K HG002.bg/metadata
2.3M HG002.bg/qc
20G HG002.bg/seqset
28G totalThese files comprise the complete BioGraph:
-
analysis/*: Thefull_pipelinescript creates this directory and writes the finalresults.vcf.gzandresults.vcf.gz.tbihere. If run with--keep, other intermediary analysis files are stored here as well. -
seqset: This is the overlap graph of all nucleotide sequences present in this BioGraph. -
coverage/*.readmap: The readmap contains coverage, pairing, and other read-related information. -
metadata/bg_info.json: This JSON file contains the mapping of sample IDs to readmap filenames and other data.
The qc/ folder contains logs, statistics, and reports from various commands. The files that are present depend on which commands have been run. In general, log files end in .txt and statistics end in .json. The runtime of each stage run by full_pipeline is saved to timings.json. The kmer_quality_report*.html files are generated during the create step. They provide a visualization of the kmer counts, which is useful for validating the cutoff chosen by the --min-kmer-count parameter.
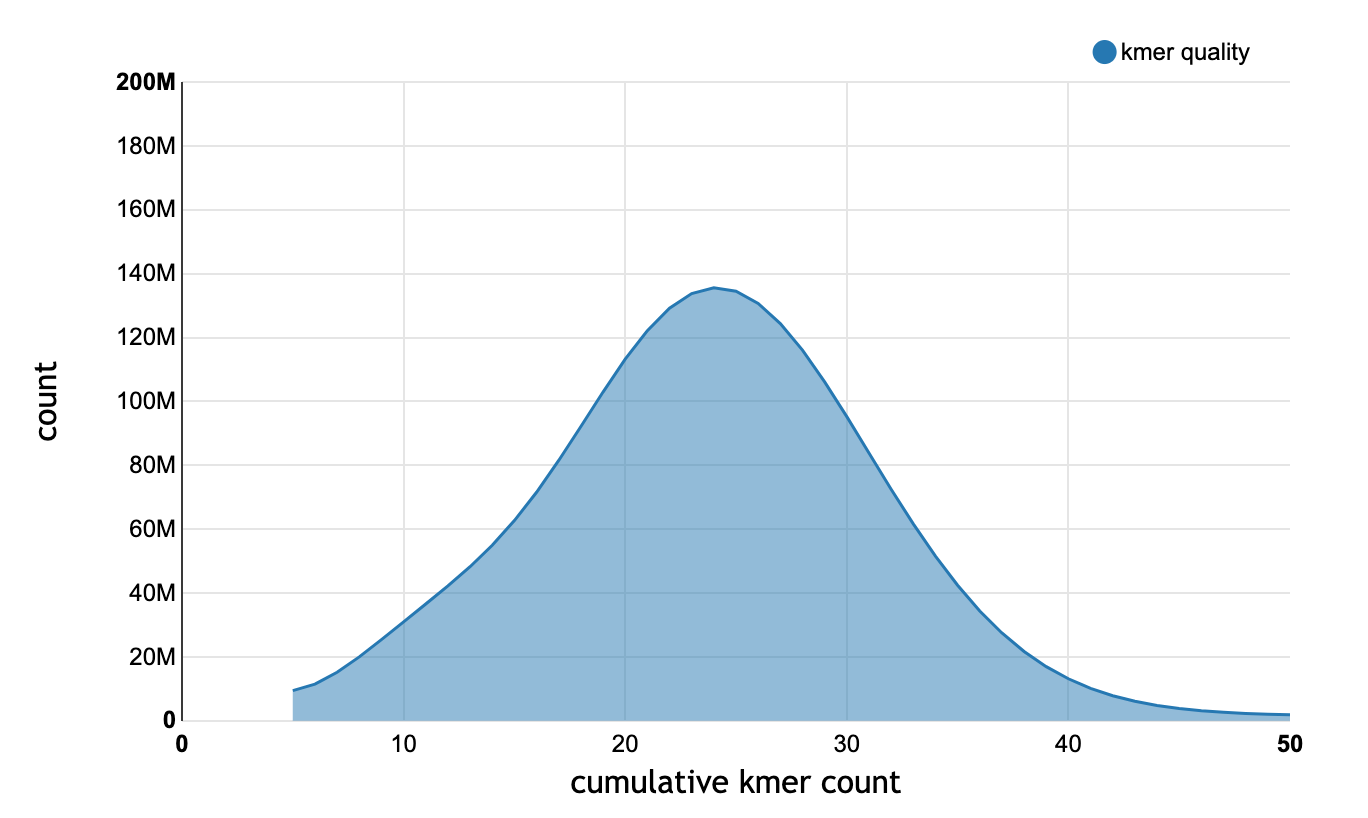
Cumulative kmer counts shown in kmer_quality_report.html.
Since all other files and directories inside the BioGraph are ignored, it can be useful to store your own QC, VCF, and other analysis results inside the BioGraph directory. Use whatever structure makes sense for your workflow to keep all of your analysis results organized and in one place.
Next: Optimizing Performance