This website contains information regarding the paper AbODE: Ab Initio Antibody Design using Conjoined ODEs.
TL;DR: We propose a generative model for antibody design using conjoined interacting neural ODEs
Please cite our work if you find it useful:
@InProceedings{pmlr-v202-verma23a,
title = {\{A\}b\{ODE\}: Ab initio antibody design using conjoined {ODE}s},
author = {Verma, Yogesh and Heinonen, Markus and Garg, Vikas},
booktitle = {Proceedings of the 40th International Conference on Machine Learning},
pages = {35037--35050},
year = {2023},
editor = {Krause, Andreas and Brunskill, Emma and Cho, Kyunghyun and Engelhardt, Barbara and Sabato, Sivan and Scarlett, Jonathan},
volume = {202},
series = {Proceedings of Machine Learning Research},
month = {23--29 Jul},
publisher = {PMLR},
pdf = {https://proceedings.mlr.press/v202/verma23a/verma23a.pdf},
url = {https://proceedings.mlr.press/v202/verma23a.html}
}
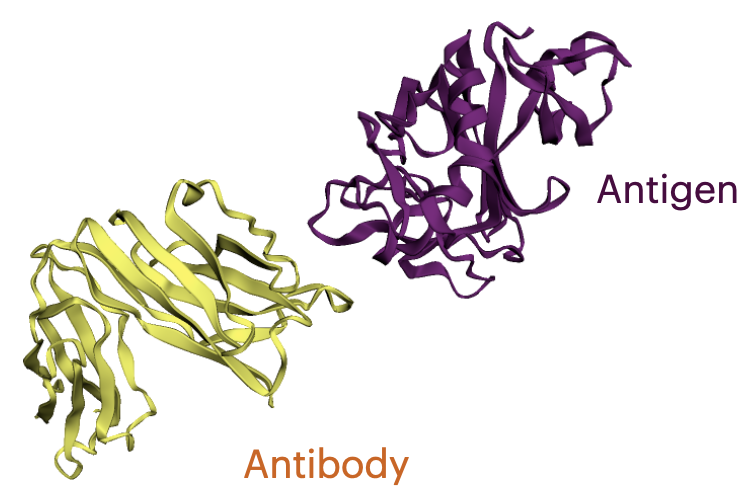
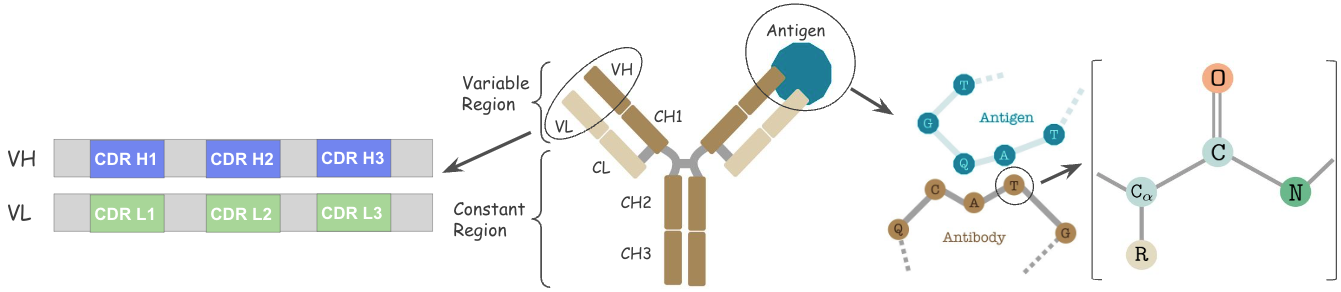
Antibodies are versatile proteins that bind to pathogens like viruses (Antigens) and stimulate a response. Each antibody recognizes a unique antigen, and the so-called Complementarity Determining Regions (CDRs) at the tip of the antibody determines this specificity.
De novo generation of new antibodies targeting specific antigens is key to accelerating vaccine discovery. However, this co-design of the amino acid sequence and the 3D structure subsumes and accentuates some central challenges from multiple tasks, including protein folding (sequence to structure), inverse folding (structure to sequence), and docking (binding). Moreover, there is a combinatorial search space of over
Neural ODEs have been widely applied to graphs, such as GRAND, PDE-GCN, etc., providing a general way to create novel embedding methods. An ODE defines the dynamics of node updates as,
Where $$\mathbf{z}{i}$$ are node features, $$f\theta$$ is parametrized by a NN, and the model essentially learns a differential vector field that guides to create of expressive embeddings of nodes.
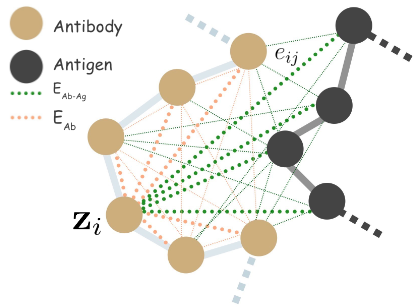
We define antigen-antibody complex as a 3D graph
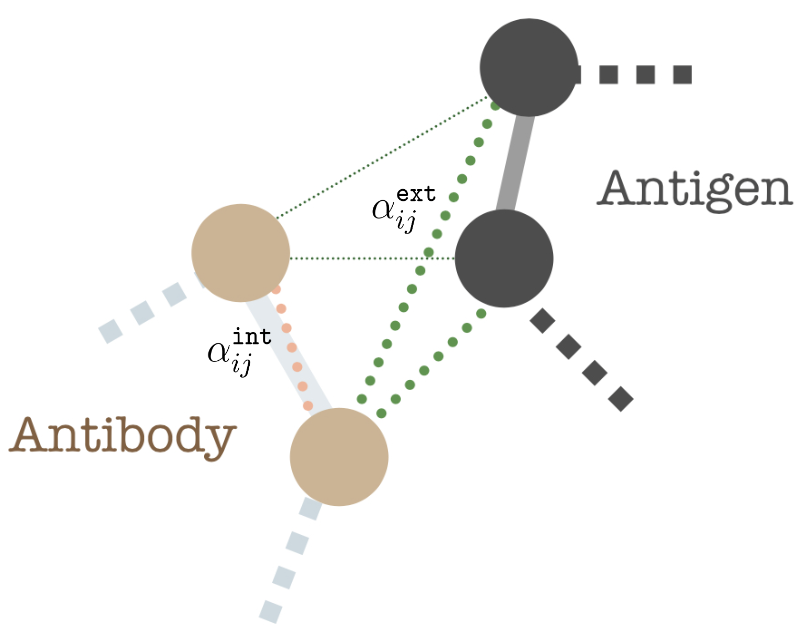
We define full connected heterogeneous edges between antibody
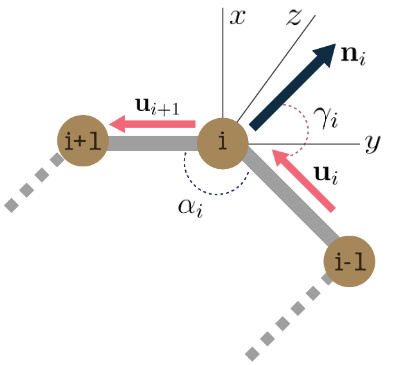
We also represent each residue by the cartesian 3D coordinates of its three backbone atoms
We model inter-antibody-antigen and intra-antibody interactions with a joint 3D graph over the antigen and the antibody using edge features,
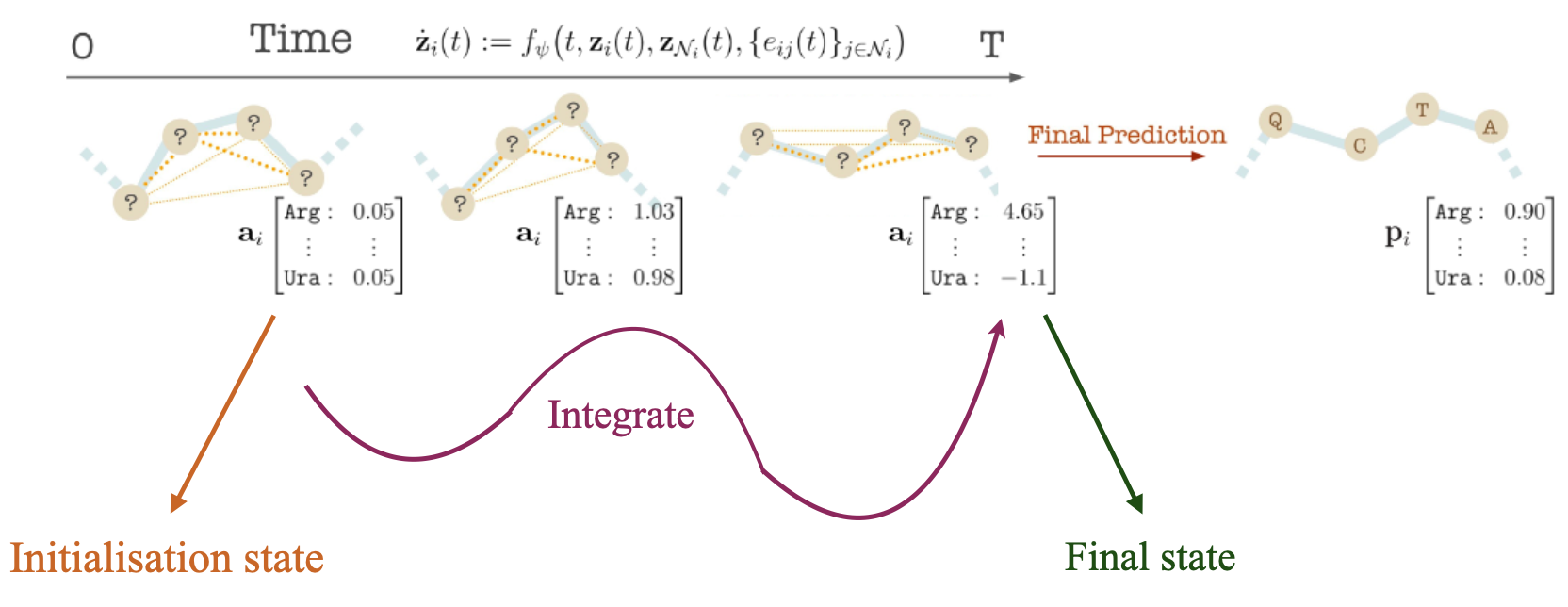
We model the distribution of antibody-antigen complexes by ODE over
We begin by assuming an ODE system $${\mathbf{z}{i}(t)}$$ over time $$t \in \mathrm{R}{+}$$, where node the time evolution of node
Collecting all, we get a system of conjoined ODEs, which can be solved using ODEsolvers.
We capture the interactions between the antigen and antibody residues with graph attention, as,
We optimize our model jointly with loss consisting of two components: one for the sequence and another for the structure, as,
where,
The angle loss is defined using negative von-mises log-likelihood and radii loss using using negative gaussian log-likelihood as,
Given the antibody or antigen-antibody complex, we generate an antibody sequence and the corresponding structure by solving the system of ODEs for time T to obtain
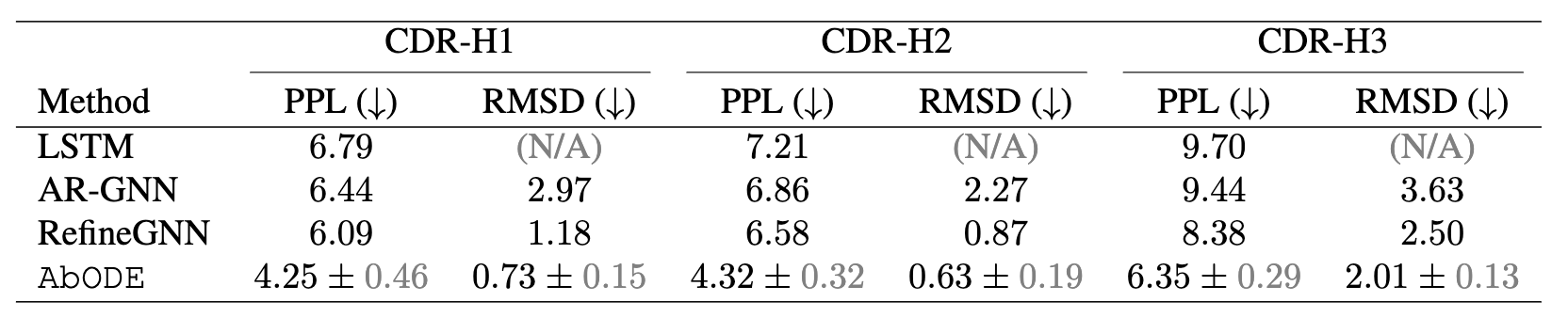
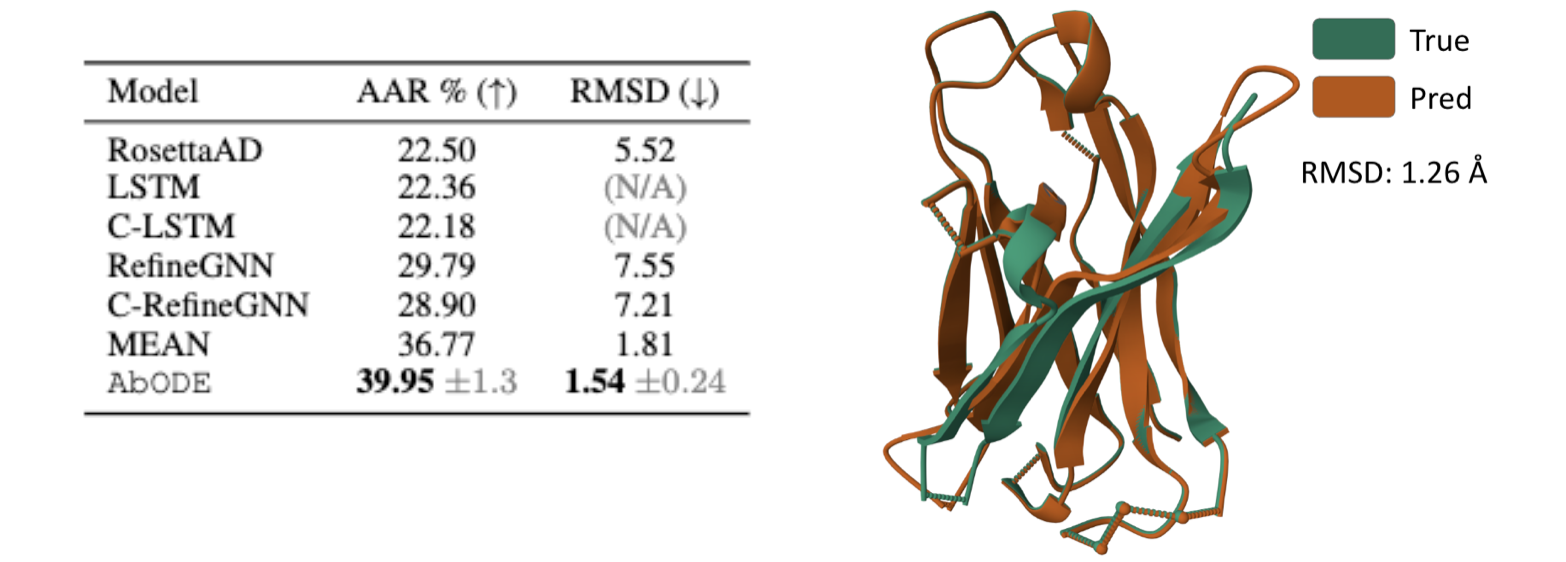
The task is to generate antibody sequence and structure without any external conditioning, where we used PPL : Perplexity (Exponential of negative log-likelihood) and RMSD: Root Mean Square Deviation by Kabsch Algorithm as our metrics.
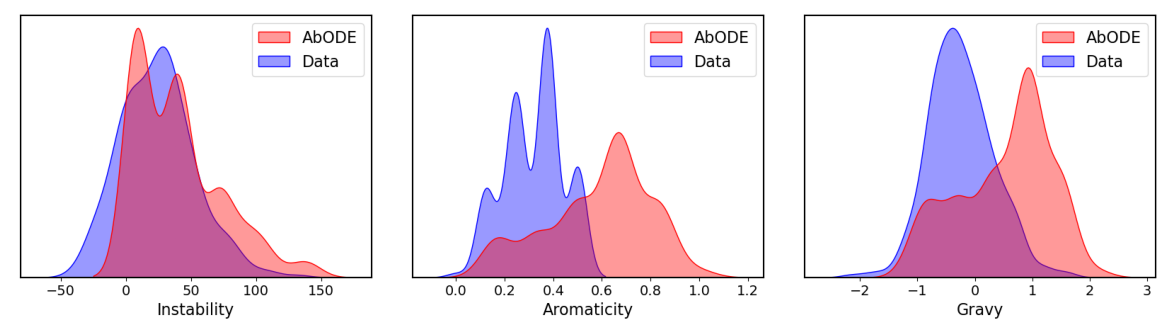
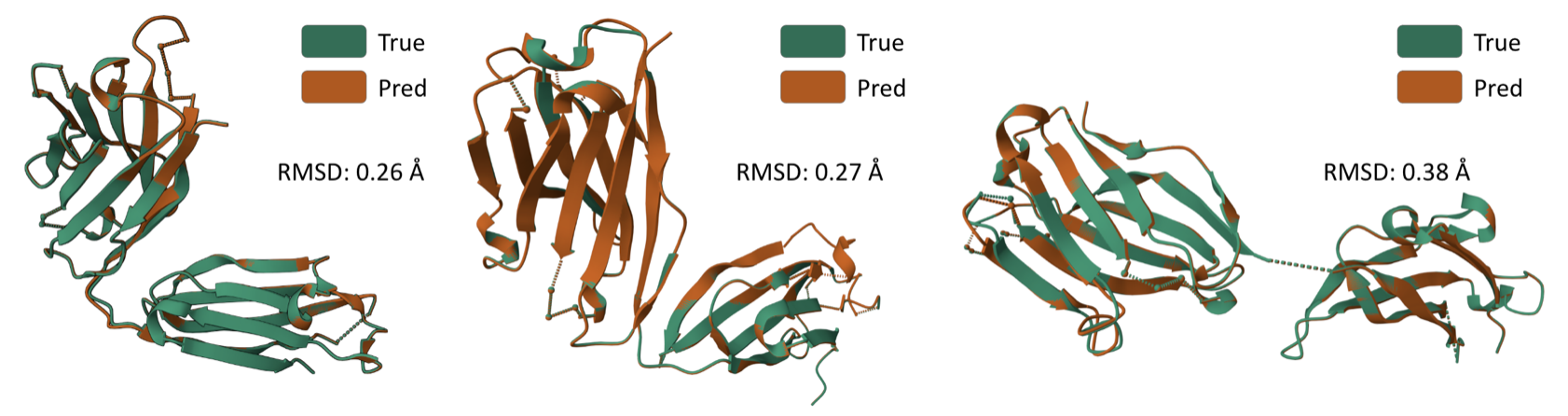
Some generated structures via **$$\texttt{AbODE}$$** are also shown above. We visually evaluate the generated structures via out method via properties distribution. We utilize kernel density estimation of these distributions to visualize these distributions. We use- Gravy: The Gravy value is calculated by adding the hydropathy value for each residue and dividing it by the sequence length.
- Instability: The Instability index predicts regional instability of dipeptides that occur more frequently in unstable proteins when compared to stable proteins
- Aromaticity: It calculates the aromaticity value of a protein, which is simply the relative frequency of Phe+Trp+Tyr
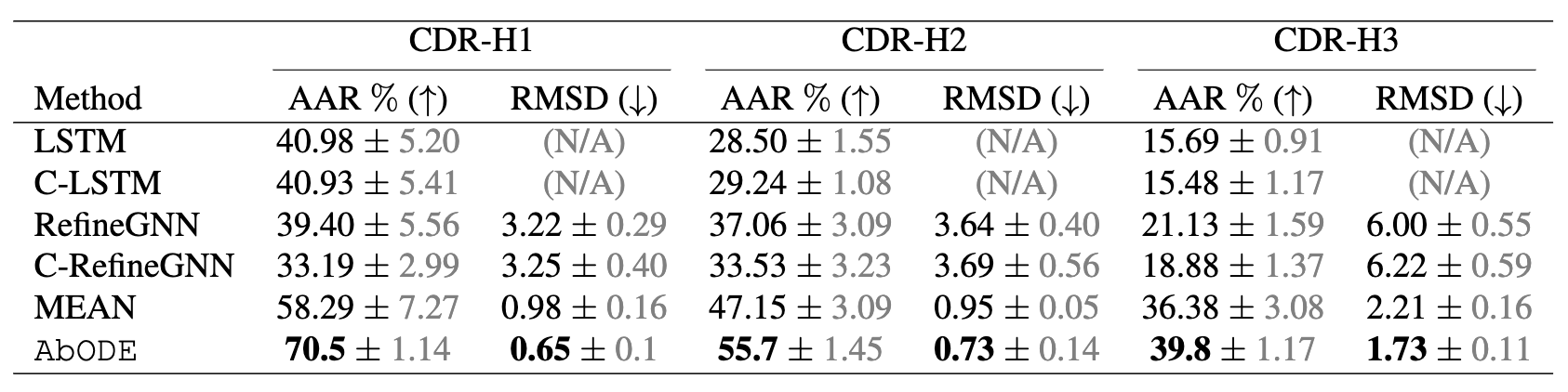
The task is to generate antibody sequence and structure with an external conditioning i.e. the Antigen, where we used AAR: Amino Acid Recovery rate, defined as the overlapping rate between the predicted 1D sequences and the ground truth and RMSD as our metrics.
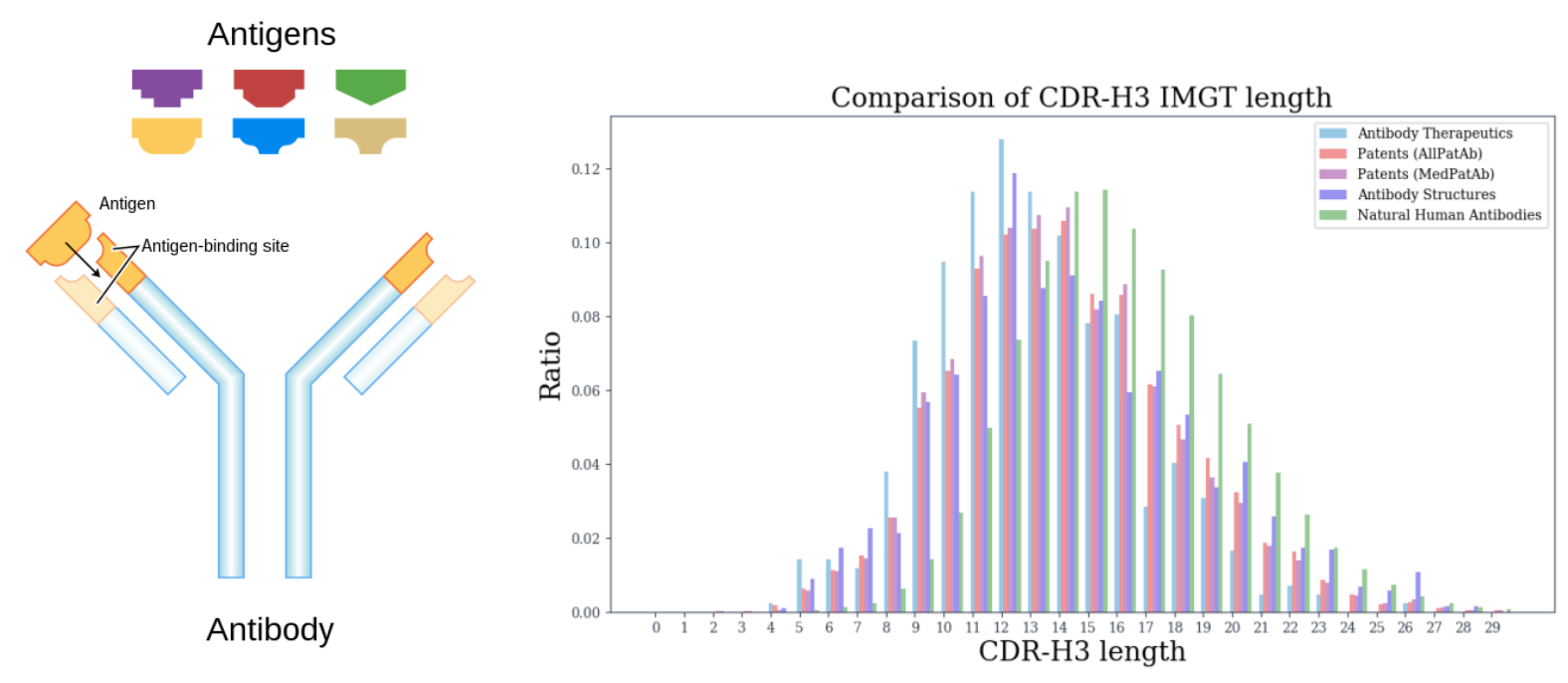
Design CDR-H3 that binds to a given antigen, evaluated on 60 diverse complexes selected by RabD
- We propose AbODE, which models the antibody-antigen complex as a joint graph, and via a system of coupled residue-specific ODEs.
- AbODE can incorporate conditional contextual and spatial information in ODEs tailored for Antibody design.
- Iterative refinement graph neural network for antibody sequence-structure co-design, Jin et al., ICLR 2022
- Independent SE(3)-Equivariant Models for End-to-End Rigid Docking, Ganea et al., ICLR 2022
- Modular Flows: DIfferential Molecular Generation, Verma et al., NeurIPS 2022
- Conditional Antibody Design as 3D Equivariant Graph Translation, Kong et al., ICLR 2023
- Generative Models for Graph-based Protein Design, Ingraham et al., NeurIPS 2019
- Neural Ordinary Differential Equations, Chen et al., NeurIPS 2018