-
Notifications
You must be signed in to change notification settings - Fork 8
How to browse multi sample VCF data in IGV
Mitch Bekritsky edited this page Oct 17, 2018
·
3 revisions
The genome VCF from the 150 Polaris 1 Diversity Cohort samples have been aggregated by the GVCF Genotyper app into one multi-sample VCF file.
This pages explains how to browse this file in the following flavours of IGV:
You can download the Desktop version of IGV from the IGV website.
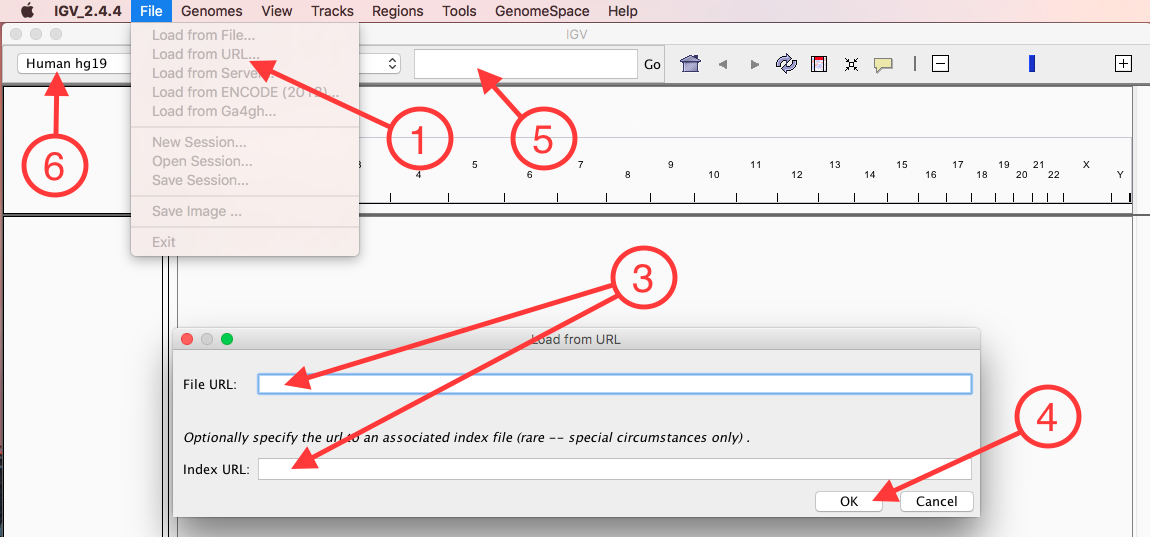
Launch IGV and (also see image below):
- Select File->Load from URL...
- Retrieve the S3 URL of two files: merged.vcf.gz and merged.vcf.gz.tbi, located in GVCF_Genotyper's appResult.
This page explains how to do so. - Back in IGV, fill in both URLs:
- File URL: S3 URL of merged.vcf.gz
- Index URL: S3 URL of merged.vcf.gz.tbi
- Confirm
- If needed: Select a region small enough for IGV to show the variants (try gene "BEST2")
- If needed: Set the reference genome to "Human hg19"