This repository contains a sample model for Task 2 of the RARE-X: A Rare Disease Open Science Data Challenge.
This model is the containerized version of the provided Jupyter notebook. The model follows the TPOT pipeline:
- add/remove features
- impute missing values
- apply other transforms
- perform Random Forest Classifier
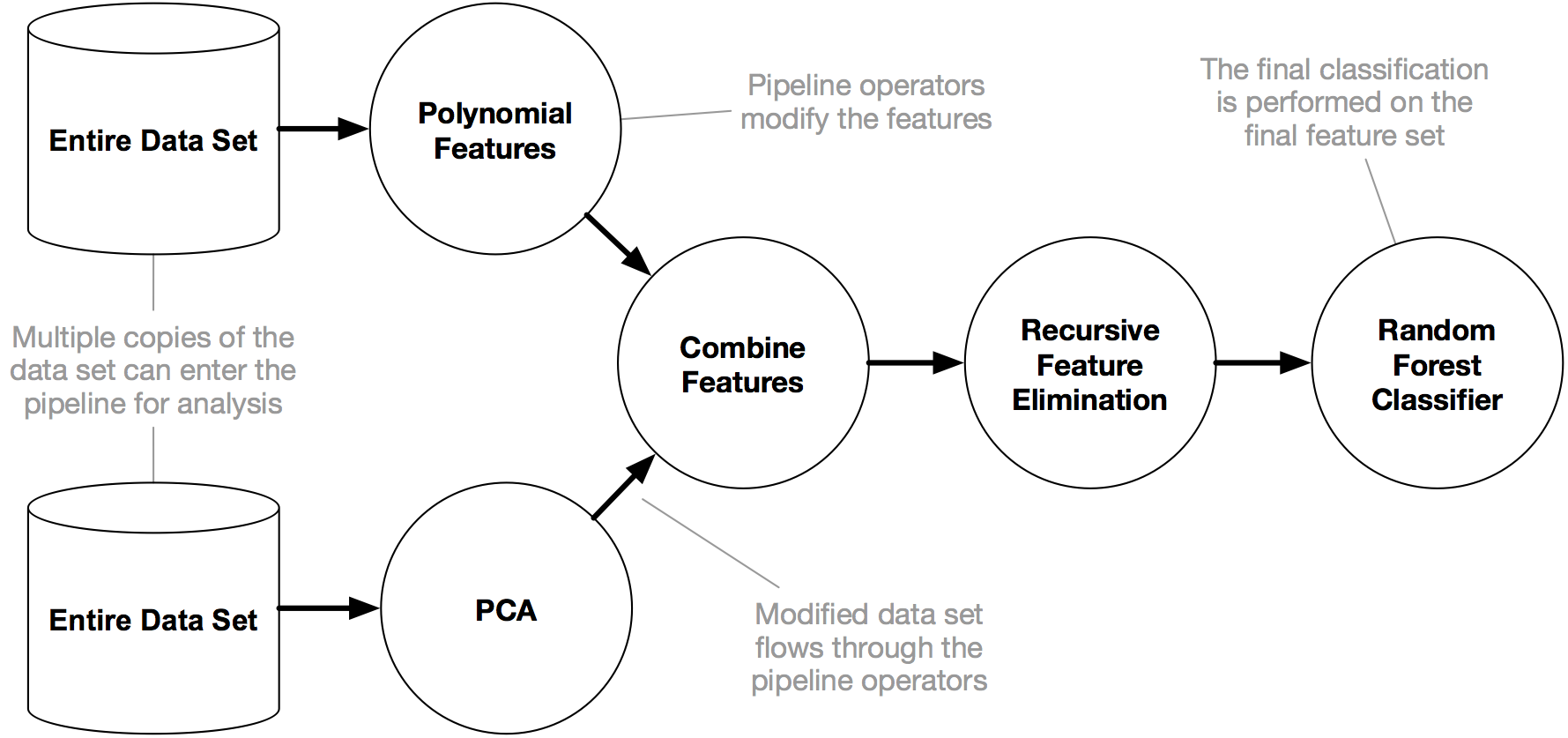 |
|---|
| Source: http://epistasislab.github.io/tpot/ |
-
Replace the TPOT pipeline with your own model!
-
Update
requirements.txtas needed. -
Dockerize the model:
docker build -t docker.synapse.org/<project id>/my-model:v1 .where:
<project id>: Synapse ID of your projectmy-model: name of your modelv1: version of your model.: filepath to the Dockerfile
Note: the Synapse submission system uses the x86-64 cpu architecture. If your machine uses a different architecture, e.g. Apple Silicon, you will need to additionally include
--platform linux/amd64into the command. -
(optional but recommended) Locally run the model to ensure it can run successfully. For this, you may use dummy_task2 and dummy_task2_test as the mounts for
/inputand/test, respectively. E.g.docker run --rm \ --network none \ --volume /path/to/dummy_task2:/input:ro \ --volume /path/to/dummy_task2_test:/test:ro \ --volume /path/to/output:/output:rw \ docker.synapse.org/<project id>/my-model:v1 -
Use
docker pushto push the model up to your project on Synapse, then submit it to the challenge.
For more information on how to submit, refer to the Submission Tutorial on the challenge site.
Author:
Jake Albrecht (@chepyle)
Contributors: