-
Notifications
You must be signed in to change notification settings - Fork 9
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Prune by distance from seed reference #290
Comments
|
I tried comparing all the different references with a few different techniques. For example, this chart shows the comparison when I calculate the Levenshtein distance to each reference. For each genotype, I calculate a median reference among all the references in that genotype. Then I display the distance from that median to all the other references. Green dots are references within the same genotype, and red dots are references in other genotypes. Genotypes 1b, 5, and 7 only have one reference each, so they are equal to their median. Most of the references within each group are within a distance of about 1500, so I'm going to try using that as a limit. Genotype 6 may cause problems, so I'll review some samples to see how they look. For comparison, here are some other techniques I used to compare the references: They all have more overlap than the first one. |
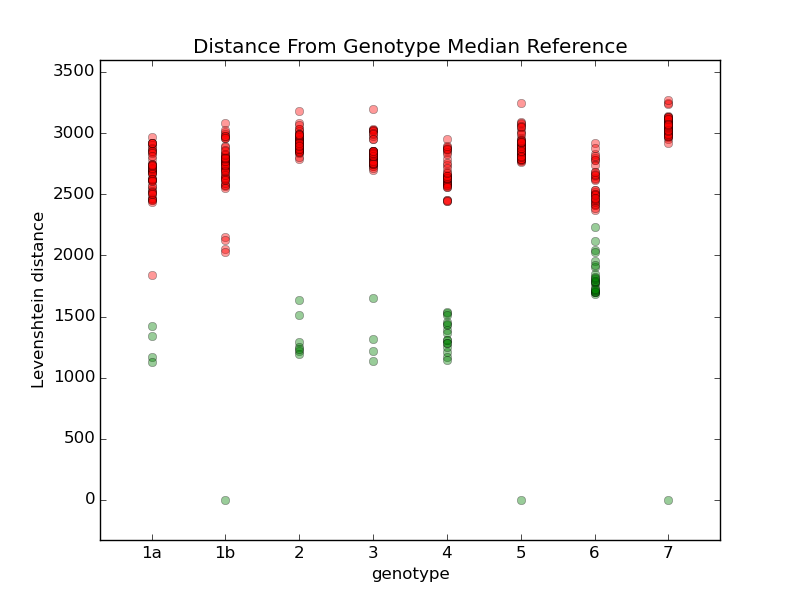
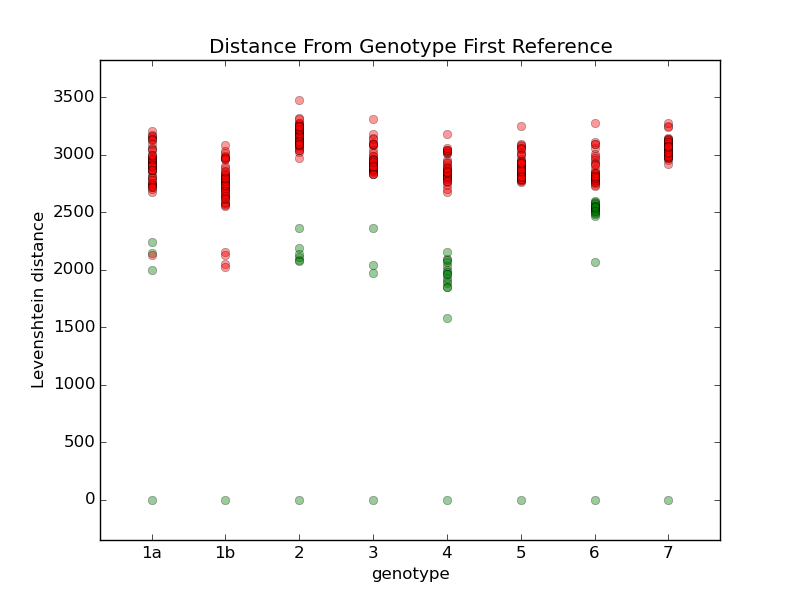
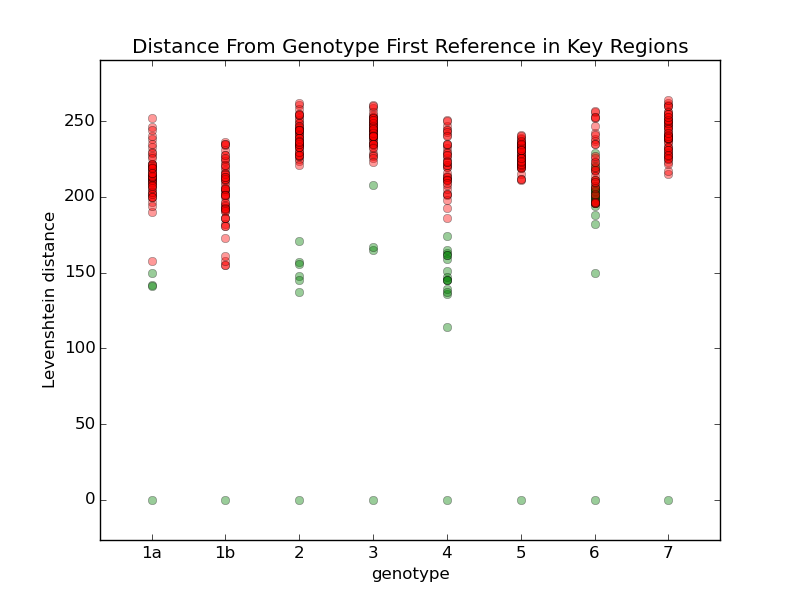
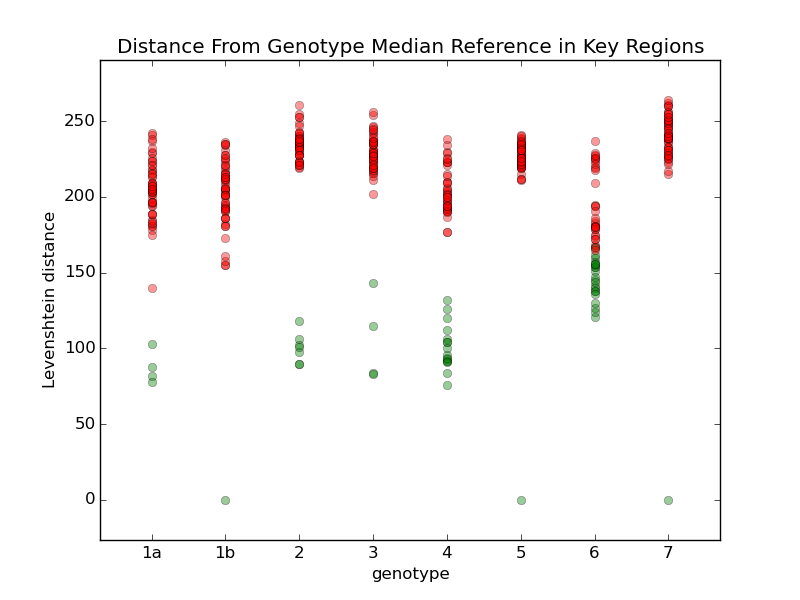
Add a step in remap that looks at how far the consensus has moved from the seed reference, and rejects any seeds that have moved more than some threshold, like 5% of the reference length. If any seeds are rejected, do another mapping with the remaining references.
Look at the Hamming or Levenshtein distance between the different references in each seed group to decide on a threshold.
As an example of a sample that moved too far from the seed reference, see samples 61673AWG1 and 61673AWG2 in the run from Mar 1 2016. The two samples are from the same extraction, but WG1 reports a mutation at NS3 155 and WG2 doesn't. We suspect that all the reads with the mutation mapped to genotype 1B and got ignored.
The text was updated successfully, but these errors were encountered: