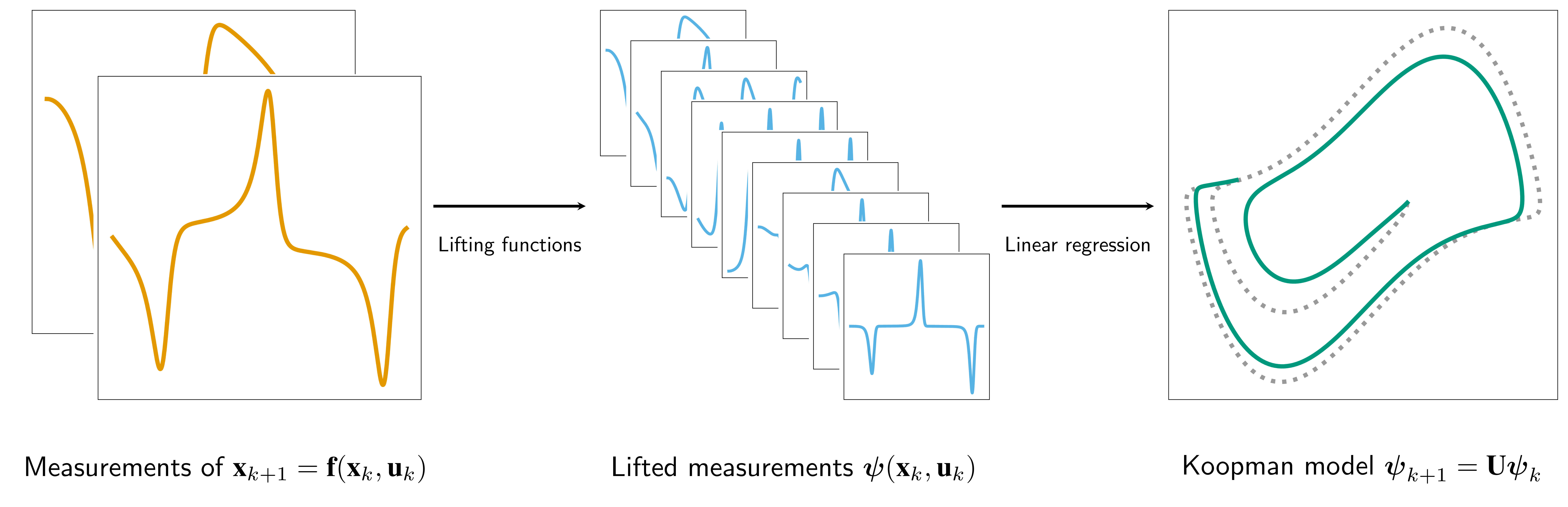
pykoop is a Koopman operator identification library written in Python. It
allows the user to specify Koopman lifting functions and regressors in order to
learn a linear model of a given system in the lifted space.
To learn more about Koopman operator theory, check out this talk or this review article.
pykoop places heavy emphasis on modular lifting function construction and
scikit-learn compatibility. The library aims to make it easy to
automatically find good lifting functions and regressor hyperparameters by
leveraging scikit-learn's existing cross-validation infrastructure.
pykoop also gracefully handles control inputs and multi-episode datasets
at every stage of the pipeline.
pykoop also includes several experimental regressors that use linear matrix
inequalities to constraint the asymptotic stability of the Koopman system, or
regularize the regression using its H-infinity norm. Check out
arXiv:2110.09658 [eess.SY] and
arXiv:2102.03613 [eess.SY] for details.
Consider Tikhonov-regularized EDMD with polynomial lifting functions applied to
mass-spring-damper data. Using pykoop, this can be implemented as:
import pykoop
from sklearn.preprocessing import MaxAbsScaler, StandardScaler
# Get example mass-spring-damper data
eg = pykoop.example_data_msd()
# Create pipeline
kp = pykoop.KoopmanPipeline(
lifting_functions=[
('ma', pykoop.SkLearnLiftingFn(MaxAbsScaler())),
('pl', pykoop.PolynomialLiftingFn(order=2)),
('ss', pykoop.SkLearnLiftingFn(StandardScaler())),
],
regressor=pykoop.Edmd(alpha=1),
)
# Fit the pipeline
kp.fit(
eg['X_train'],
n_inputs=eg['n_inputs'],
episode_feature=eg['episode_feature'],
)
# Predict using the pipeline
X_pred = kp.predict_trajectory(eg['x0_valid'], eg['u_valid'])
# Score using the pipeline
score = kp.score(eg['X_valid'])
# Plot results
kp.plot_predicted_trajectory(eg['X_valid'], plot_input=True)More examples are available in examples/, in notebooks/, or on
binder.
Most of the required classes and functions have been imported into the
pykoop namespace. The most important object is the
pykoop.KoopmanPipeline, which requires a list of lifting functions and
a regressor.
Some example lifting functions are
pykoop.PolynomialLiftingFn,pykoop.RbfLiftingFn,pykoop.DelayLiftingFn, andpykoop.BilinearInputLiftingFn.
scikit-learn preprocessors can be wrapped into lifting functions using
pykoop.SkLearnLiftingFn. States and inputs can be lifted independently
using pykoop.SplitPipeline. This is useful to avoid lifting inputs.
Some basic regressors included are
pykoop.Edmd(includes Tikhonov regularization),pykoop.Dmdc, andpykoop.Dmd.
More advanced (and experimental) LMI-based regressors are included in the
pykoop.lmi_regressors namespace. They allow for different kinds of
regularization as well as hard constraints on the Koopman operator.
You can roll your own lifting functions and regressors by inheriting from
pykoop.KoopmanLiftingFn, pykoop.EpisodeIndependentLiftingFn,
pykoop.EpisodeDependentLiftingFn, and
pykoop.KoopmanRegressor.
Some sample dynamic models are also included in the pykoop.dynamic_models
namespace.
pykoop can be installed from PyPI using
$ pip install pykoopAdditional LMI solvers can be installed using
$ pip install mosek
$ pip install cvxopt
$ pip install smcpMosek is recommended, but is nonfree and requires a license.
The library can be tested using
$ pip install -r requirements-dev.txt
$ pytestNote that pytest must be run from the repository's root directory.
To skip unit tests that require a MOSEK license, including all doctests and examples, run
$ pytest ./tests -k "not mosek"The documentation can be compiled using
$ cd doc
$ make htmlIf you want a hook to check source code formatting before allowing a commit, you can use
$ cd .git/hooks/
$ ln -s ../../.githooks/pre-commit .
$ chmod +x ./pre-commitYou will need yapf installed for this.
Other excellent Python packages for learning dynamical systems exist, summarized in the table below:
| Library | Unique features |
|---|---|
| pykoop |
|
| pykoopman |
|
| PyDMD |
|
| PySINDy |
|
If you use this software in your research, please cite it as below or see
CITATION.cff.
@software{dahdah_pykoop_2024,
title={{decargroup/pykoop}},
doi={10.5281/zenodo.5576490},
url={https://github.com/decargroup/pykoop},
publisher={Zenodo},
author={Steven Dahdah and James Richard Forbes},
version = {{v2.0.2}},
year={2024},
}This project is distributed under the MIT License, except the contents of
pykoop/_sklearn_metaestimators/, which are from the scikit-learn
project, and are distributed under the BSD-3-Clause License.