-
Notifications
You must be signed in to change notification settings - Fork 7
Indexing
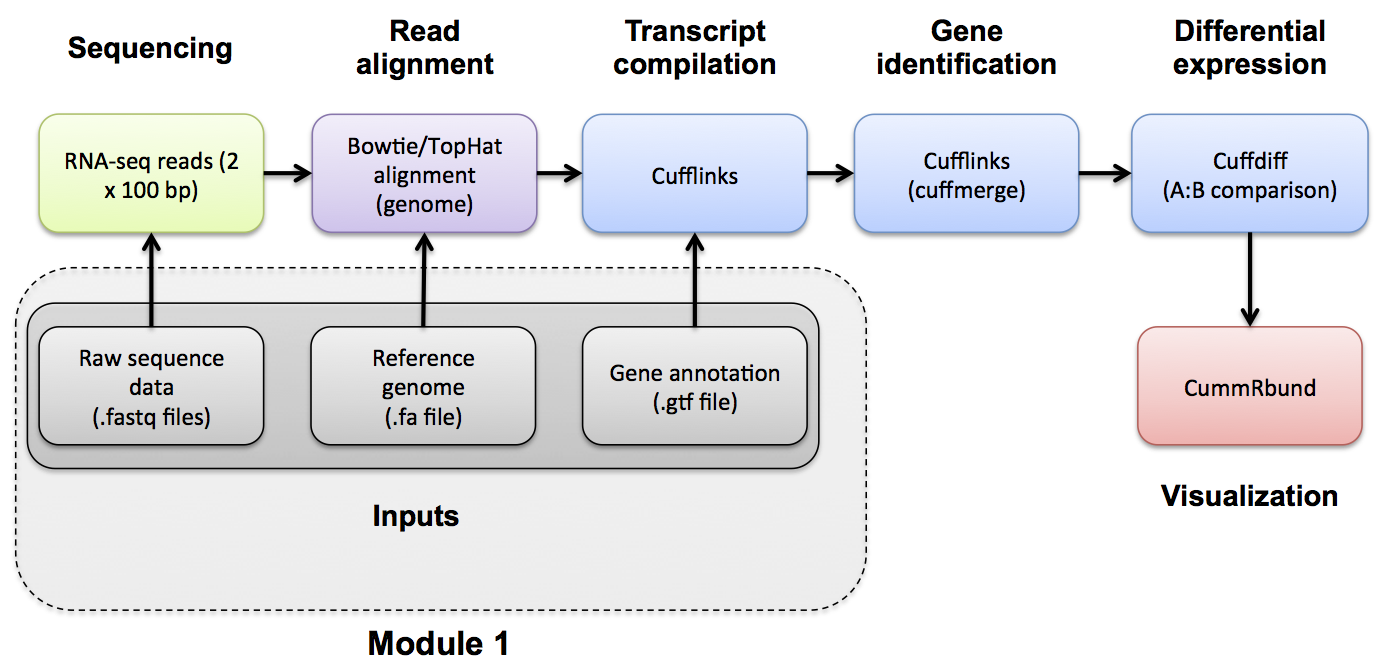
#1-iv. Indexing
###Create a bowtie2 index
Create a bowtie index for chr22 and the ERCC spike-in sequences and write it to a 'bwt' sub-directory:
cd $RNA_HOME/refs/hg19/
mkdir -p bwt/chr22_ERCC92
bowtie2-build fasta/chr22_ERCC92/chr22_ERCC92.fa bwt/chr22_ERCC92/chr22_ERCC92
cp $RNA_HOME/refs/hg19/fasta/chr22_ERCC92/*.fa $RNA_HOME/refs/hg19/bwt/chr22_ERCC92/
ls bwt/chr22_ERCC92/
To create an index for all chromosomes instead of just chr22 you would do the following:
#cd $RNA_HOME/refs/hg19/
#mkdir bwt
#bowtie2-build fasta/hg19.fa bwt/hg19
#mv $RNA_HOME/refs/hg19/fasta/hg19.fa $RNA_HOME/refs/hg19/bwt/
###OPTIONAL ALTERNATIVE - STAR indexes Create index files for use with STAR
cd $RNA_HOME/refs/hg19/
mkdir -p star/chr22_ERCC92
STAR --runMode genomeGenerate --genomeDir $RNA_HOME/refs/hg19/star/chr22_ERCC92 --genomeFastaFiles $RNA_HOME/refs/hg19/fasta/chr22_ERCC92/chr22_ERCC92.fa --runThreadN 8 --sjdbGTFfile $RNA_HOME/refs/hg19/genes/genes_chr22_ERCC92.gtf --sjdbOverhang 100
####OPTIONAL ALTERNATIVE - HISAT2 indexes Create reference and splice site index files for use with HISAT2
cd $RNA_HOME/refs/hg19/
mkdir -p hisat2/chr22_ERCC92
hisat2-build $RNA_HOME/refs/hg19/fasta/chr22_ERCC92/chr22_ERCC92.fa $RNA_HOME/refs/hg19/hisat2/chr22_ERCC92/chr22_ERCC92
extract_splice_sites.py $RNA_HOME/refs/hg19/genes/genes_chr22_ERCC92.gtf > $RNA_HOME/refs/hg19/hisat2/chr22_ERCC92/splicesites.txt
| Previous Section | This Section | Next Section | |:-------------------------------:|:---------------------------:|:----------------------------:| | Annotations | Indexing | Data |
##Note: The current version of this tutorial is now at www.rnaseq.wiki
Table of Contents
Module 0: Authors | Citation | Syntax | Intro to AWS | Log into AWS | Unix | Environment | Resources
Module 1: Installation | Reference Genomes | Annotations | Indexing | Data | Data QC
Module 2: Adapter Trim | Alignment | IGV | Alignment Visualization | Alignment QC
Module 3: Expression | Differential Expression | DE Visualization
Module 4: Ref Guided | De novo | Merging | Differential Splicing | Splicing Visualization
Module 5: Kallisto
Appendix: Abbreviations | Lectures | Practical Exercise Solutions | Integrated Assignment | Proposed Improvements | AWS Setup