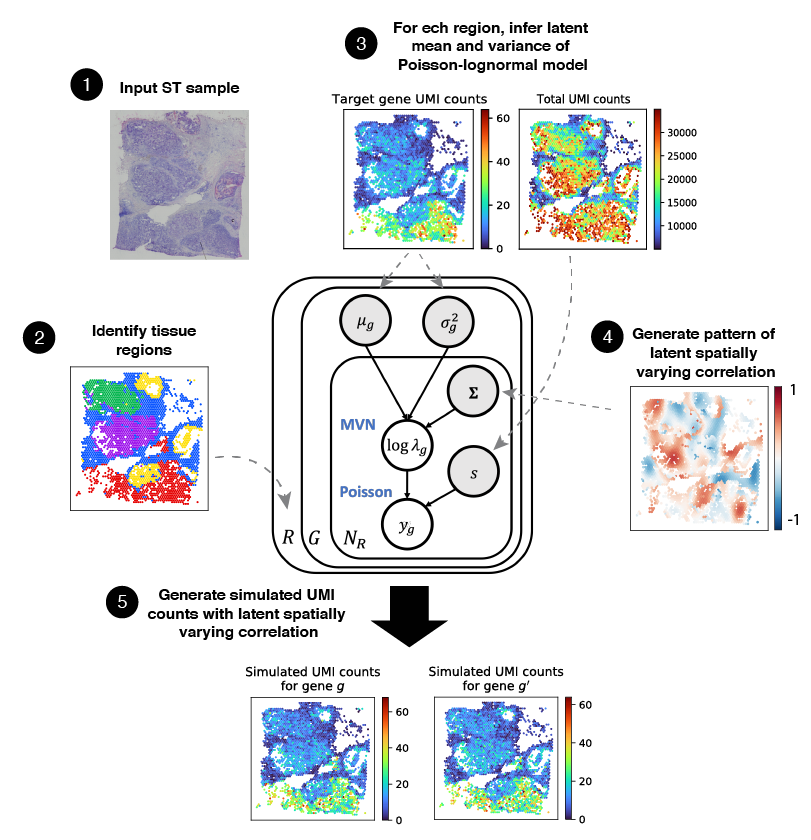
This package simulates spatial transcriptomics data with spatially varying correlation structure. To do so, it uses a Poisson-lognormal model with parameters inferred from an input dataset.
Here is a schematic overview of the simulation framework:
To install SpatialCorr-sim using Pip, run the following command:
pip install spatialcorr-sim
The API manual can be found at https://spatialcorr-sim.readthedocs.io/en/latest/.
For a tutorial on how to simulate a spatial transcriptomics dataset, please see the Jupyter notebook, tutorial/SpatialCorr_sim_tutorial.ipynb.